In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases
- PMID: 22761581
- PMCID: PMC3386304
- DOI: 10.1371/journal.pgen.1002750
In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases
Abstract
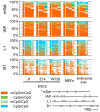
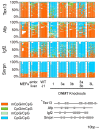
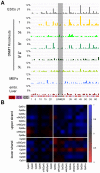
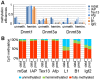
The enzymatic control of the setting and maintenance of symmetric and non-symmetric DNA methylation patterns in a particular genome context is not well understood. Here, we describe a comprehensive analysis of DNA methylation patterns generated by high resolution sequencing of hairpin-bisulfite amplicons of selected single copy genes and repetitive elements (LINE1, B1, IAP-LTR-retrotransposons, and major satellites). The analysis unambiguously identifies a substantial amount of regional incomplete methylation maintenance, i.e. hemimethylated CpG positions, with variant degrees among cell types. Moreover, non-CpG cytosine methylation is confined to ESCs and exclusively catalysed by Dnmt3a and Dnmt3b. This sequence position-, cell type-, and region-dependent non-CpG methylation is strongly linked to neighboring CpG methylation and requires the presence of Dnmt3L. The generation of a comprehensive data set of 146,000 CpG dyads was used to apply and develop parameter estimated hidden Markov models (HMM) to calculate the relative contribution of DNA methyltransferases (Dnmts) for de novo and maintenance DNA methylation. The comparative modelling included wild-type ESCs and mutant ESCs deficient for Dnmt1, Dnmt3a, Dnmt3b, or Dnmt3a/3b, respectively. The HMM analysis identifies a considerable de novo methylation activity for Dnmt1 at certain repetitive elements and single copy sequences. Dnmt3a and Dnmt3b contribute de novo function. However, both enzymes are also essential to maintain symmetrical CpG methylation at distinct repetitive and single copy sequences in ESCs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Comment in
-
DNA methylation: in vivo enzyme action.Nat Rev Genet. 2012 Jul 18;13(8):520. doi: 10.1038/nrg3284. Nat Rev Genet. 2012. PMID: 22805707 No abstract available.
References
-
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. - PubMed
-
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. - PubMed
-
- Bird A, Taggart M, Frommer M, Miller OJ, Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40:91–99. - PubMed
-
- Gama-Sosa MA, Midgett RM, Slagel VA, Githens S, Kuo KC, et al. Tissue-specific differences in DNA methylation in various mammals. Biochim Biophys Acta. 1983;740:212–219. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

