Identification of kinases regulating prostate cancer cell growth using an RNAi phenotypic screen
- PMID: 22761715
- PMCID: PMC3384611
- DOI: 10.1371/journal.pone.0038950
Identification of kinases regulating prostate cancer cell growth using an RNAi phenotypic screen
Abstract
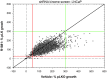
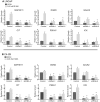
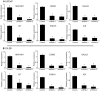
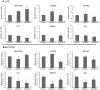
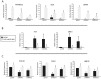
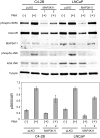
As prostate cancer progresses to castration-resistant disease, there is an increase in signal transduction activity. Most castration-resistant prostate tumors continue to express the androgen receptor (AR) as well as androgen-responsive genes, despite the near absence of circulating androgen in these patients. The AR is regulated not only by its cognate steroid hormone, but also by interactions with a constellation of co-regulatory and signaling molecules. Thus, the elevated signaling activity that occurs during progression to castration resistance can affect prostate cancer cell growth either through the AR or independent of the AR. In order to identify signaling pathways that regulate prostate cancer cell growth, we screened a panel of shRNAs targeting 673 human kinases against LNCaP prostate cancer cells grown in the presence and absence of hormone. The screen identified multiple shRNA clones against known and novel gene targets that regulate prostate cancer cell growth. Based on the magnitude of effect on growth, we selected six kinases for further study: MAP3K11, DGKD, ICK, CIT, GALK2, and PSKH1. Knockdown of these kinases decreased cell growth in both androgen-dependent and castration-resistant prostate cancer cells. However, these kinases had different effects on basal or androgen-induced transcriptional activity of AR target genes. MAP3K11 knockdown most consistently altered transcription of AR target genes, suggesting that MAP3K11 affected its growth inhibitory effect by modulating the AR transcriptional program. Consistent with MAP3K11 acting on the AR, knockdown of MAP3K11 inhibited AR Ser 650 phosphorylation, further supporting stress kinase regulation of AR phosphorylation. This study demonstrates the applicability of lentiviral-based shRNA for conducting phenotypic screens and identifies MAP3K11, DGKD, ICK, CIT, GALK2, and PSKH1 as regulators of prostate cancer cell growth. The thorough evaluation of these kinase targets will pave the way for developing more effective treatments for castration-resistant prostate cancer.
Conflict of interest statement
Figures
References
-
- Gioeli D. Signal transduction in prostate cancer progression. Clin Sci (Lond) 2005;108:293–308. - PubMed
-
- Cunha GR, Ricke W, Thomson A, Marker PC, Risbridger G, et al. Hormonal, cellular, and molecular regulation of normal and neoplastic prostatic development. J Steroid Biochem Mol Biol. 2004;92:221–36. - PubMed
-
- Burnstein KL. Regulation of androgen receptor levels: implications for prostate cancer progression and therapy. J Cell Biochem. 2005;95:657–69. - PubMed
-
- Perez CA. Cancer of the Prostate. In: Cancer. Principles and Practice of Oncology. Philadelphia: J.B. Lippincott Company. 1985. pp. 929–964.
-
- Gioeli DG. The promise of novel androgen receptor antagonists. Cell Cycle. 2010;9:440–1. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

