Detection of ligation products of DNA linkers with 5'-OH ends by denaturing PAGE silver stain
- PMID: 22761747
- PMCID: PMC3384673
- DOI: 10.1371/journal.pone.0039251
Detection of ligation products of DNA linkers with 5'-OH ends by denaturing PAGE silver stain
Abstract
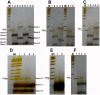
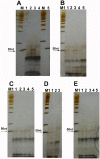
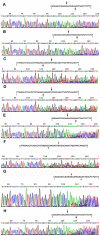
To explore if DNA linkers with 5'-hydroxyl (OH) ends could be joined by commercial T4 and E. coli DNA ligase, these linkers were synthesized by using the solid-phase phosphoramidite method and joined by using commercial T4 and E. coli DNA ligases. The ligation products were detected by using denaturing PAGE silver stain and PCR method. About 0.5-1% of linkers A-B and E-F, and 0.13-0.5% of linkers C-D could be joined by T4 DNA ligases. About 0.25-0.77% of linkers A-B and E-F, and 0.06-0.39% of linkers C-D could be joined by E. coli DNA ligases. A 1-base deletion (-G) and a 5-base deletion (-GGAGC) could be found at the ligation junctions of the linkers. But about 80% of the ligation products purified with a PCR product purification kit did not contain these base deletions, meaning that some linkers had been correctly joined by T4 and E. coli DNA ligases. In addition, about 0.025-0.1% of oligo 11 could be phosphorylated by commercial T4 DNA ligase. The phosphorylation products could be increased when the phosphorylation reaction was extended from 1 hr to 2 hrs. We speculated that perhaps the linkers with 5'-OH ends could be joined by T4 or E. coli DNA ligase in 2 different manners: (i) about 0.025-0.1% of linkers could be phosphorylated by commercial T4 DNA ligase, and then these phosphorylated linkers could be joined to the 3'-OH ends of other linkers; and (ii) the linkers could delete one or more nucleotide(s) at their 5'-ends and thereby generated some 5'-phosphate ends, and then these 5'-phosphate ends could be joined to the 3'-OH ends of other linkers at a low efficiency. Our findings may probably indicate that some DNA nicks with 5'-OH ends can be joined by commercial T4 or E. coli DNA ligase even in the absence of PNK.
Conflict of interest statement
Figures






References
-
- Lehman IR. DNA ligase: structure, mechanism, and function. Science. 1974;186:790–797. - PubMed
-
- Timson DJ, Singleton MR, Wigley DB. DNA ligases in the repair and replication of DNA. Mutat Res. 2000;460:301–318. - PubMed
-
- Wilkinson A, Day J, Bowater R. Bacterial DNA ligases. Mol Microbiol. 2001;40:1241–1248. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

