Biochemistry and cell biology of tau protein in neurofibrillary degeneration
- PMID: 22762014
- PMCID: PMC3385935
- DOI: 10.1101/cshperspect.a006247
Biochemistry and cell biology of tau protein in neurofibrillary degeneration
Abstract
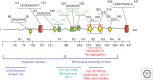
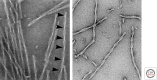
Tau represents the subunit protein of one of the major hallmarks of Alzheimer disease (AD), the neurofibrillary tangles, and is therefore of major interest as an indicator of disease mechanisms. Many of the unusual properties of Tau can be explained by its nature as a natively unfolded protein. Examples are the large number of structural conformations and biochemical modifications (phosphorylation, proteolysis, glycosylation, and others), the multitude of interaction partners (mainly microtubules, but also other cytoskeletal proteins, kinases, and phosphatases, motor proteins, chaperones, and membrane proteins). The pathological aggregation of Tau is counterintuitive, given its high solubility, but can be rationalized by short hydrophobic motifs forming β structures. The aggregation of Tau is toxic in cell and animal models, but can be reversed by suppressing expression or by aggregation inhibitors. This review summarizes some of the structural, biochemical, and cell biological properties of Tau and Tau fibers. Further aspects of Tau as a diagnostic marker and therapeutic target, its involvement in other Tau-based diseases, and its histopathology are covered by other chapters in this volume.
Figures
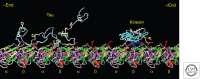


References
-
- Aamodt EJ, Williams RC Jr 1984. Microtubule-associated proteins connect microtubules and neurofilaments in vitro. Biochemistry 23: 6023–6031 - PubMed
-
- Ackmann M, Wiech H, Mandelkow E 2000. Nonsaturable binding indicates clustering of tau on the microtubule surface in a paired helical filament-like conformation. J Biol Chem 275: 30335–30343 - PubMed
-
- Aguzzi A, Rajendran L 2009. The transcellular spread of cytosolic amyloids, prions, and prionoids. Neuron 64: 783–790 - PubMed
-
- Ahn JS, Radhakrishnan ML, Mapelli M, Choi S, Tidor B, Cuny GD, Musacchio A, Yeh LA, Kosik KS 2005. Defining Cdk5 ligand chemical space with small molecule inhibitors of tau phosphorylation. Chem Biol 12: 811–823 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
