HIV-1 assembly, budding, and maturation
- PMID: 22762019
- PMCID: PMC3385941
- DOI: 10.1101/cshperspect.a006924
HIV-1 assembly, budding, and maturation
Erratum in
- Cold Spring Harb Perspect Med. 2012 Aug;2(8). doi: 10.1101/cshperspect.a015420
Abstract
A defining property of retroviruses is their ability to assemble into particles that can leave producer cells and spread infection to susceptible cells and hosts. Virion morphogenesis can be divided into three stages: assembly, wherein the virion is created and essential components are packaged; budding, wherein the virion crosses the plasma membrane and obtains its lipid envelope; and maturation, wherein the virion changes structure and becomes infectious. All of these stages are coordinated by the Gag polyprotein and its proteolytic maturation products, which function as the major structural proteins of the virus. Here, we review our current understanding of the mechanisms of HIV-1 assembly, budding, and maturation, starting with a general overview and then providing detailed descriptions of each of the different stages of virion morphogenesis.
Figures





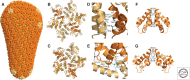
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
