Genomics and bioinformatics of Parkinson's disease
- PMID: 22762024
- PMCID: PMC3385936
- DOI: 10.1101/cshperspect.a009449
Genomics and bioinformatics of Parkinson's disease
Abstract
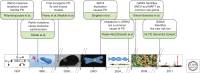
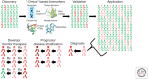
Within the last two decades, genomics and bioinformatics have profoundly impacted our understanding of the molecular mechanisms of Parkinson's disease (PD). From the description of the first PD gene in 1997 until today, we have witnessed the emergence of new technologies that have revolutionized our concepts to identify genetic mechanisms implicated in human health and disease. Driven by the publication of the human genome sequence and followed by the description of detailed maps for common genetic variability, novel applications to rapidly scrutinize the entire genome in a systematic, cost-effective manner have become a reality. As a consequence, about 30 genetic loci have been unequivocally linked to the pathogenesis of PD highlighting essential molecular pathways underlying this common disorder. Herein we discuss how neurogenomics and bioinformatics are applied to dissect the nature of this complex disease with the overall aim of developing rational therapeutic interventions.
Figures


Similar articles
-
Exploring Parkinson's through the Lens of Genomics and Bioinformatics.Cold Spring Harb Perspect Med. 2025 Feb 10:a041621. doi: 10.1101/cshperspect.a041621. Online ahead of print. Cold Spring Harb Perspect Med. 2025. PMID: 39929729
-
Role of genomics in translational research for Parkinson's disease.Biochem Biophys Res Commun. 2014 Sep 19;452(2):226-35. doi: 10.1016/j.bbrc.2014.06.028. Epub 2014 Jun 17. Biochem Biophys Res Commun. 2014. PMID: 24950403 Review.
-
Contribution of genomics and proteomics in understanding the role of modifying factors in Parkinson's disease.Indian J Biochem Biophys. 2006 Apr;43(2):69-81. Indian J Biochem Biophys. 2006. PMID: 16955754 Review.
-
Functional genomics elucidates regulatory mechanisms of Parkinson's disease-associated variants.BMC Med. 2022 Feb 16;20(1):68. doi: 10.1186/s12916-022-02264-w. BMC Med. 2022. PMID: 35168626 Free PMC article.
-
Establishing gene regulatory networks from Parkinson's disease risk loci.Brain. 2022 Jul 29;145(7):2422-2435. doi: 10.1093/brain/awac022. Brain. 2022. PMID: 35094046 Free PMC article.
Cited by
-
Proteomics in immunity and herpes simplex encephalitis.Expert Rev Proteomics. 2014 Feb;11(1):21-9. doi: 10.1586/14789450.2014.864954. Epub 2013 Dec 18. Expert Rev Proteomics. 2014. PMID: 24351021 Free PMC article. Review.
-
Biomarkers of Parkinson's disease: present and future.Metabolism. 2015 Mar;64(3 Suppl 1):S40-6. doi: 10.1016/j.metabol.2014.10.030. Epub 2014 Oct 31. Metabolism. 2015. PMID: 25510818 Free PMC article. Review.
-
Analysis of total RNA as a potential biomarker of Parkinson's disease in silico.Int J Immunopathol Pharmacol. 2025 Jan-Dec;39:3946320241297738. doi: 10.1177/03946320241297738. Int J Immunopathol Pharmacol. 2025. PMID: 39819073 Free PMC article.
-
Parkinson's disease.Subcell Biochem. 2012;65:389-455. doi: 10.1007/978-94-007-5416-4_16. Subcell Biochem. 2012. PMID: 23225012 Free PMC article. Review.
-
Regional analysis and genetic association of nigrostriatal degeneration in Lewy body disease.Mov Disord. 2017 Nov;32(11):1584-1593. doi: 10.1002/mds.27184. Epub 2017 Sep 26. Mov Disord. 2017. PMID: 28949048 Free PMC article.
References
-
- Andre F, Job B, Dessen P, Tordai A, Michiels S, Liedtke C, Richon C, Yan K, Wang B, Vassal G, et al. 2009. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res 15: 441–451 - PubMed
-
- Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M, et al. 2003. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 299: 256–259 - PubMed
-
- Braak H, Del Tredici K, Bratzke H, Hamm-Clement J, Sandmann-Keil D, Rub U 2002. Staging of the intracerebral inclusion body pathology associated with idiopathic Parkinson's disease (preclinical and clinical stages). J Neurol 249: III/1–5 - PubMed
-
- Braak H, Del Tredici K, Rub U, de Vos RA, Jansen Steur EN, Braak E 2003a. Staging of brain pathology related to sporadic Parkinson's disease. Neurobiol Aging 24: 197–211 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
