Functional roles of protein splicing factors
- PMID: 22762203
- PMCID: PMC3392075
- DOI: 10.1042/BSR20120007
Functional roles of protein splicing factors
Abstract
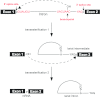
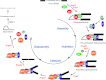
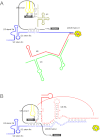
RNA splicing is one of the fundamental processes in gene expression in eukaryotes. Splicing of pre-mRNA is catalysed by a large ribonucleoprotein complex called the spliceosome, which consists of five small nuclear RNAs and numerous protein factors. The spliceosome is a highly dynamic structure, assembled by sequential binding and release of the small nuclear RNAs and protein factors. DExD/H-box RNA helicases are required to mediate structural changes in the spliceosome at various steps in the assembly pathway and have also been implicated in the fidelity control of the splicing reaction. Other proteins also play key roles in mediating the progression of the spliceosome pathway. In this review, we discuss the functional roles of the protein factors involved in the spliceosome pathway primarily from studies in the yeast system.
Figures

References
-
- Brody E., Abelson J. The ‘spliceosome’: yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science. 1985;228:963–967. - PubMed
-
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985;42:355–367. - PubMed
-
- Grabowski P. J., Seiler S. R., Sharp P. A. A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell. 1985;42:345–353. - PubMed
-
- Wahl M. C., Will C. L., Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. - PubMed
-
- Brow D. A. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

