P-bodies and stress granules: possible roles in the control of translation and mRNA degradation
- PMID: 22763747
- PMCID: PMC3428773
- DOI: 10.1101/cshperspect.a012286
P-bodies and stress granules: possible roles in the control of translation and mRNA degradation
Abstract
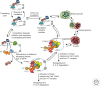
The control of translation and mRNA degradation is important in the regulation of eukaryotic gene expression. In general, translation and steps in the major pathway of mRNA decay are in competition with each other. mRNAs that are not engaged in translation can aggregate into cytoplasmic mRNP granules referred to as processing bodies (P-bodies) and stress granules, which are related to mRNP particles that control translation in early development and neurons. Analyses of P-bodies and stress granules suggest a dynamic process, referred to as the mRNA Cycle, wherein mRNPs can move between polysomes, P-bodies and stress granules although the functional roles of mRNP assembly into higher order structures remain poorly understood. In this article, we review what is known about the coupling of translation and mRNA degradation, the properties of P-bodies and stress granules, and how assembly of mRNPs into larger structures might influence cellular function.
Figures


References
-
- Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M 2008. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol 10: 1324–1332 - PubMed
-
- Arkov A, Wang JY, Ramos A, Lehmann R 2006. The role of Tudor domains in germline development and polar granule architecture. Development 133: 4053–4062 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
