Nature and function of insulator protein binding sites in the Drosophila genome
- PMID: 22767387
- PMCID: PMC3483548
- DOI: 10.1101/gr.138156.112
Nature and function of insulator protein binding sites in the Drosophila genome
Erratum in
- Genome Res. 2013 Feb;23(2):409
Abstract
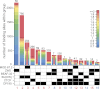
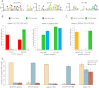
Chromatin insulator elements and associated proteins have been proposed to partition eukaryotic genomes into sets of independently regulated domains. Here we test this hypothesis by quantitative genome-wide analysis of insulator protein binding to Drosophila chromatin. We find distinct combinatorial binding of insulator proteins to different classes of sites and uncover a novel type of insulator element that binds CP190 but not any other known insulator proteins. Functional characterization of different classes of binding sites indicates that only a small fraction act as robust insulators in standard enhancer-blocking assays. We show that insulators restrict the spreading of the H3K27me3 mark but only at a small number of Polycomb target regions and only to prevent repressive histone methylation within adjacent genes that are already transcriptionally inactive. RNAi knockdown of insulator proteins in cultured cells does not lead to major alterations in genome expression. Taken together, these observations argue against the concept of a genome partitioned by specialized boundary elements and suggest that insulators are reserved for specific regulation of selected genes.
Figures



References
-
- Baniahmad A, Steiner C, Köhne AC, Renkawitz R 1990. Modular structure of a chicken lysozyme silencer: Involvement of an unusual thyroid hormone receptor binding site. Cell 61: 505–514 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
