Deconjugation of Nedd8 from Cul1 is directly regulated by Skp1-F-box and substrate, and the COP9 signalosome inhibits deneddylated SCF by a noncatalytic mechanism
- PMID: 22767593
- PMCID: PMC3436198
- DOI: 10.1074/jbc.M112.352484
Deconjugation of Nedd8 from Cul1 is directly regulated by Skp1-F-box and substrate, and the COP9 signalosome inhibits deneddylated SCF by a noncatalytic mechanism
Abstract
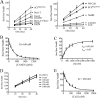
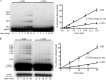
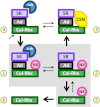
COP9 signalosome (CSN) mediates deconjugation of the ubiquitin-like protein Nedd8 from the cullin subunits of SCF and other cullin-RING ubiquitin ligases (CRLs). This process is essential to maintain the proper activity of CRLs in cells. Here, we report a detailed kinetic characterization of CSN-mediated deconjugation of Nedd8 from SCF. CSN is an efficient enzyme, with a k(cat) of ~1 s(-1) and K(m) for neddylated Cul1-Rbx1 of ~200 nm, yielding a k(cat)/K(m) near the anticipated diffusion-controlled limit. Assembly with an F-box-Skp1 complex markedly inhibited deneddylation, although the magnitude varied considerably, with Fbw7-Skp1 inhibiting by ~5-fold but Skp2-Cks1-Skp1 by only ~15%. Deneddylation of both SCF(Fbw7) and SCF(Skp2-Cks1) was further inhibited ~2.5-fold by the addition of substrate. Combined, the inhibition by Fbw7-Skp1 plus its substrate cyclin E was greater than 10-fold. Unexpectedly, our results also uncover significant product inhibition by deconjugated Cul1, which results from the ability of Cul1 to bind tightly to CSN. Reciprocally, CSN inhibits the ubiquitin ligase activity of deneddylated Cul1. We propose a model in which assembled CRL complexes engaged with substrate are normally refractory to deneddylation. Upon consumption of substrate and subsequent deneddylation, CSN can remain stably bound to the CRL and hold it in low state of reduced activity.
Figures



Similar articles
-
Regulation of neddylation and deneddylation of cullin1 in SCFSkp2 ubiquitin ligase by F-box protein and substrate.Proc Natl Acad Sci U S A. 2006 Aug 1;103(31):11515-20. doi: 10.1073/pnas.0603921103. Epub 2006 Jul 21. Proc Natl Acad Sci U S A. 2006. PMID: 16861300 Free PMC article.
-
HSC70 coordinates COP9 signalosome and SCF ubiquitin ligase activity to enable a prompt stress response.EMBO Rep. 2025 Mar;26(5):1344-1366. doi: 10.1038/s44319-025-00376-x. Epub 2025 Feb 6. EMBO Rep. 2025. PMID: 39915298 Free PMC article.
-
CAND1 enhances deneddylation of CUL1 by COP9 signalosome.Biochem Biophys Res Commun. 2005 Sep 2;334(3):867-74. doi: 10.1016/j.bbrc.2005.06.188. Biochem Biophys Res Commun. 2005. PMID: 16036220
-
Protection of cullin-RING E3 ligases by CSN-UBP12.Trends Cell Biol. 2006 Jul;16(7):362-9. doi: 10.1016/j.tcb.2006.05.001. Epub 2006 Jun 9. Trends Cell Biol. 2006. PMID: 16762551 Review.
-
Deregulation of the COP9 signalosome-cullin-RING ubiquitin-ligase pathway: mechanisms and roles in urological cancers.Int J Biochem Cell Biol. 2013 Jul;45(7):1327-37. doi: 10.1016/j.biocel.2013.03.023. Epub 2013 Apr 10. Int J Biochem Cell Biol. 2013. PMID: 23583660 Review.
Cited by
-
The cyclomodulin cycle inhibiting factor (CIF) alters cullin neddylation dynamics.J Biol Chem. 2013 May 24;288(21):14716-26. doi: 10.1074/jbc.M112.448258. Epub 2013 Apr 15. J Biol Chem. 2013. PMID: 23589306 Free PMC article.
-
CSN5A Subunit of COP9 Signalosome Is Required for Resetting Transcriptional Stress Memory after Recurrent Heat Stress in Arabidopsis.Biomolecules. 2021 Apr 30;11(5):668. doi: 10.3390/biom11050668. Biomolecules. 2021. PMID: 33946149 Free PMC article.
-
Plant deubiquitinases: from structure and activity to biological functions.Plant Cell Rep. 2023 Mar;42(3):469-486. doi: 10.1007/s00299-022-02962-y. Epub 2022 Dec 26. Plant Cell Rep. 2023. PMID: 36567335 Review.
-
Overexpression of COP9 signalosome subunits, CSN7A and CSN7B, exerts different effects on adipogenic differentiation.FEBS Open Bio. 2016 Oct 11;6(11):1102-1112. doi: 10.1002/2211-5463.12129. eCollection 2016 Nov. FEBS Open Bio. 2016. PMID: 27833851 Free PMC article.
-
Hepatic deficiency of COP9 signalosome subunit 8 induces ubiquitin-proteasome system impairment and Bim-mediated apoptosis in murine livers.PLoS One. 2013 Jul 1;8(7):e67793. doi: 10.1371/journal.pone.0067793. Print 2013. PLoS One. 2013. PMID: 23840878 Free PMC article.
References
-
- Petroski M. D., Deshaies R. J. (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 - PubMed
-
- Deshaies R. J., Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 - PubMed
-
- Soucy T. A., Smith P. G., Milhollen M. A., Berger A. J., Gavin J. M., Adhikari S., Brownell J. E., Burke K. E., Cardin D. P., Critchley S., Cullis C. A., Doucette A., Garnsey J. J., Gaulin J. L., Gershman R. E., Lublinsky A. R., McDonald A., Mizutani H., Narayanan U., Olhava E. J., Peluso S., Rezaei M., Sintchak M. D., Talreja T., Thomas M. P., Traore T., Vyskocil S., Weatherhead G. S., Yu J., Zhang J., Dick L. R., Claiborne C. F., Rolfe M., Bolen J. B., Langston S. P. (2009) An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 - PubMed
-
- Cardozo T., Pagano M. (2004) The SCF ubiquitin ligase. Insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

