Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology
- PMID: 22768112
- PMCID: PMC3387205
- DOI: 10.1371/journal.pone.0039720
Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology
Abstract
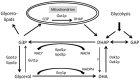
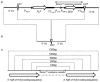
Targeted gene replacement to generate knock-outs and knock-ins is a commonly used method to study the function of unknown genes. In the methylotrophic yeast Pichia pastoris, the importance of specific gene targeting has increased since the genome sequencing projects of the most commonly used strains have been accomplished, but rapid progress in the field has been impeded by inefficient mechanisms for accurate integration. To improve gene targeting efficiency in P. pastoris, we identified and deleted the P. pastoris KU70 homologue. We observed a substantial increase in the targeting efficiency using the two commonly known and used integration loci HIS4 and ADE1, reaching over 90% targeting efficiencies with only 250-bp flanking homologous DNA. Although the ku70 deletion strain was noted to be more sensitive to UV rays than the corresponding wild-type strain, no lethality, severe growth retardation or loss of gene copy numbers could be detected during repetitive rounds of cultivation and induction of heterologous protein production. Furthermore, we demonstrated the use of the ku70 deletion strain for fast and simple screening of genes in the search of new auxotrophic markers by targeting dihydroxyacetone synthase and glycerol kinase genes. Precise knock-out strains for the well-known P. pastoris AOX1, ARG4 and HIS4 genes and a whole series of expression vectors were generated based on the wild-type platform strain, providing a broad spectrum of precise tools for both intracellular and secreted production of heterologous proteins utilizing various selection markers and integration strategies for targeted or random integration of single and multiple genes. The simplicity of targeted integration in the ku70 deletion strain will further support protein production strain generation and synthetic biology using P. pastoris strains as platform hosts.
Conflict of interest statement
Figures
References
-
- Lin Cereghino GP, Lin Cereghino J, Ilgen C, Cregg JM. Production of recombinant proteins in fermenter cultures of the yeast Pichia pastoris. Curr Opin Biotechnol. 2002;13:329–332. - PubMed
-
- De Schutter K, Lin Y-C, Tiels P, Van Hecke A, Glinka S, et al. Genome sequence of the recombinant protein production host Pichia pastoris. Nat Biotechnol. 2009;27:561–566. - PubMed
-
- Küberl A, Schneider J, Thallinger GG, Anderl I, Wibberg D, et al. High-quality genome sequence of Pichia pastoris CBS7435. J Biotechnol. 2011;154:312–320. - PubMed
-
- Pastwa E, Blasiak J. Non-homologous DNA end joining. Acta Biochim Pol (Engl Transl) 2003;50:891–908. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

