Molecular epidemiology of HIV-1 subtypes in India: origin and evolutionary history of the predominant subtype C
- PMID: 22768132
- PMCID: PMC3387228
- DOI: 10.1371/journal.pone.0039819
Molecular epidemiology of HIV-1 subtypes in India: origin and evolutionary history of the predominant subtype C
Abstract
Background: India has the third largest HIV-1 epidemic with 2.4 million infected individuals. Molecular epidemiological analysis has identified the predominance of HIV-1 subtype C (HIV-1C). However, the previous reports have been limited by sample size, and uneven geographical distribution. The introduction of HIV-1C in India remains uncertain due to this lack of structured studies. To fill the gap, we characterised the distribution pattern of HIV-1 subtypes in India based on data collection from nationwide clinical cohorts between 2007 and 2011. We also reconstructed the time to the most recent common ancestor (tMRCA) of the predominant HIV-1C strains.
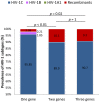
Methodology/principal findings: Blood samples were collected from 168 HIV-1 seropositive subjects from 7 different states. HIV-1 subtypes were determined using two or three genes, gag, pol, and env using several methods. Bayesian coalescent-based approach was used to reconstruct the time of introduction and population growth patterns of the Indian HIV-1C. For the first time, a high prevalence (10%) of unique recombinant forms (BC and A1C) was observed when two or three genes were used instead of one gene (p<0.01; p = 0.02, respectively). The tMRCA of Indian HIV-1C was estimated using the three viral genes, ranged from 1967 (gag) to 1974 (env). Pol-gene analysis was considered to provide the most reliable estimate [1971, (95% CI: 1965-1976)]. The population growth pattern revealed an initial slow growth phase in the mid-1970s, an exponential phase through the 1980s, and a stationary phase since the early 1990s.
Conclusions/significance: The Indian HIV-1C epidemic originated around 40 years ago from a single or few genetically related African lineages, and since then largely evolved independently. The effective population size in the country has been broadly stable since the 1990s. The evolving viral epidemic, as indicated by the increase of recombinant strains, warrants a need for continued molecular surveillance to guide efficient disease intervention strategies.
Conflict of interest statement
Figures



References
-
- Simoes EA, Babu PG, John TJ, Nirmala S, Solomon S, et al. Evidence for HTLV-III infection in prostitutes in Tamil Nadu (India). Indian J Med Res. 1987;85:335–338. - PubMed
-
- Neogi U, Sood V, Banerjee S, Ghosh N, Verma S, et al. Global HIV-1 molecular epidemiology with special reference to genetic analysis of HIV-1 subtypes circulating in North India: functional and pathogenic implications of genetic variation. Indian J Exp Biol. 2009;47:424–431. - PubMed
-
- Kandathil AJ, Ramalingam S, Kannangai R, David S, Sridharan G. Molecular epidemiology of HIV. Indian J Med Res. 2005;121:333–344. - PubMed
-
- Neogi U, Sahoo PN, Arumugam K, Sonnerborg A, DeCosta A, et al. Higher prevalence of predicted X4-tropic strains in perinatally-infected older children with HIV-1 subtype C in India. J Acquir Immune Defic Syndr. 2012;59:347–353. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

