Identification of enriched driver gene alterations in subgroups of non-small cell lung cancer patients based on histology and smoking status
- PMID: 22768234
- PMCID: PMC3387024
- DOI: 10.1371/journal.pone.0040109
Identification of enriched driver gene alterations in subgroups of non-small cell lung cancer patients based on histology and smoking status
Abstract
Background: Appropriate patient selection is needed for targeted therapies that are efficacious only in patients with specific genetic alterations. We aimed to define subgroups of patients with candidate driver genes in patients with non-small cell lung cancer.
Methods: Patients with primary lung cancer who underwent clinical genetic tests at Guangdong General Hospital were enrolled. Driver genes were detected by sequencing, high-resolution melt analysis, qPCR, or multiple PCR and RACE methods.
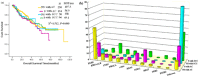
Results: 524 patients were enrolled in this study, and the differences in driver gene alterations among subgroups were analyzed based on histology and smoking status. In a subgroup of non-smokers with adenocarcinoma, EGFR was the most frequently altered gene, with a mutation rate of 49.8%, followed by EML4-ALK (9.3%), PTEN (9.1%), PIK3CA (5.2%), c-Met (4.8%), KRAS (4.5%), STK11 (2.7%), and BRAF (1.9%). The three most frequently altered genes in a subgroup of smokers with adenocarcinoma were EGFR (22.0%), STK11 (19.0%), and KRAS (12.0%). We only found EGFR (8.0%), c-Met (2.8%), and PIK3CA (2.6%) alterations in the non-smoker with squamous cell carcinoma (SCC) subgroup. PTEN (16.1%), STK11 (8.3%), and PIK3CA (7.2%) were the three most frequently enriched genes in smokers with SCC. DDR2 and FGFR2 only presented in smokers with SCC (4.4% and 2.2%, respectively). Among these four subgroups, the differences in EGFR, KRAS, and PTEN mutations were statistically significant.
Conclusion: The distinct features of driver gene alterations in different subgroups based on histology and smoking status were helpful in defining patients for future clinical trials that target these genes. This study also suggests that we may consider patients with infrequent alterations of driver genes as having rare or orphan diseases that should be managed with special molecularly targeted therapies.
Conflict of interest statement
Figures


References
-
- Mitsudomi T. Advances in target therapy for lung cancer. Jpn J Clin Oncol. 2010;40:101–106. - PubMed
-
- Bronte G, Rizzo S, La Paglia L, Adamo V, Siragusa S, et al. Driver mutations and differential sensitivity to targeted therapies: a new approach to the treatment of lung adenocarcinoma. Cancer Treat Rev. 2010;36:S21–29. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

