Characterization of plasmids in a human clinical strain of Lactococcus garvieae
- PMID: 22768237
- PMCID: PMC3387028
- DOI: 10.1371/journal.pone.0040119
Characterization of plasmids in a human clinical strain of Lactococcus garvieae
Abstract
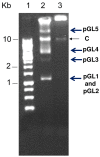
The present work describes the molecular characterization of five circular plasmids found in the human clinical strain Lactococcus garvieae 21881. The plasmids were designated pGL1-pGL5, with molecular sizes of 4,536 bp, 4,572 bp, 12,948 bp, 14,006 bp and 68,798 bp, respectively. Based on detailed sequence analysis, some of these plasmids appear to be mosaics composed of DNA obtained by modular exchange between different species of lactic acid bacteria. Based on sequence data and the derived presence of certain genes and proteins, the plasmid pGL2 appears to replicate via a rolling-circle mechanism, while the other four plasmids appear to belong to the group of lactococcal theta-type replicons. The plasmids pGL1, pGL2 and pGL5 encode putative proteins related with bacteriocin synthesis and bacteriocin secretion and immunity. The plasmid pGL5 harbors genes (txn, orf5 and orf25) encoding proteins that could be considered putative virulence factors. The gene txn encodes a protein with an enzymatic domain corresponding to the family actin-ADP-ribosyltransferases toxins, which are known to play a key role in pathogenesis of a variety of bacterial pathogens. The genes orf5 and orf25 encode two putative surface proteins containing the cell wall-sorting motif LPXTG, with mucin-binding and collagen-binding protein domains, respectively. These proteins could be involved in the adherence of L. garvieae to mucus from the intestine, facilitating further interaction with intestinal epithelial cells and to collagenous tissues such as the collagen-rich heart valves. To our knowledge, this is the first report on the characterization of plasmids in a human clinical strain of this pathogen.
Conflict of interest statement
Figures








References
-
- Fefer JJ, Ratzan KR, Sharp SE, Saiz E. Lactococcus garvieae endocarditis: report of a case and review of the literature. Diagn Microbiol Infect Dis. 1998;32:127–130. - PubMed
-
- Vinh DC, Nichol KA, Rand F, Embil JM. Native-valve bacterial endocarditis caused by Lactococcus garvieae. Diagn Microbiol Infect Dis. 2006;56:91–94. - PubMed
-
- Li WK, Chen YS, Wann SR, Liu YC, Tsai HC. Lactococcus garvieae endocarditis with initial presentation of acute cerebral infarction in a healthy immunocompetent man. Intern Med. 2008;12:1143–1146. - PubMed
-
- Mofredj A, Baraka D, Kloeti G, Dumont JL. Lactococcus garvieae septicemia with liver abscess in an immunosuppressed patient. Am J Med. 2000;109:513–514. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

