Translational control of cell division by Elongator
- PMID: 22768388
- PMCID: PMC3388810
- DOI: 10.1016/j.celrep.2012.04.001
Translational control of cell division by Elongator
Abstract
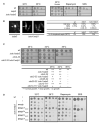
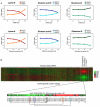
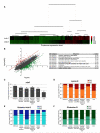
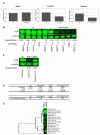
Elongator is required for the synthesis of the mcm(5)s(2) modification found on tRNAs recognizing AA-ending codons. In order to obtain a global picture of the role of Elongator in translation, we used reverse protein arrays to screen the fission yeast proteome for translation defects. Unexpectedly, this revealed that Elongator inactivation mainly affected three specific functional groups including proteins implicated in cell division. The absence of Elongator results in a delay in mitosis onset and cytokinesis defects. We demonstrate that the kinase Cdr2, which is a central regulator of mitosis and cytokinesis, is under translational control by Elongator due to the Lysine codon usage bias of the cdr2 coding sequence. These findings uncover a mechanism by which the codon usage, coupled to tRNA modifications, fundamentally contributes to gene expression and cellular functions.
Figures
References
-
- Agris PF, Vendeix FA, Graham WD. tRNA’s wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007;366:1–13. - PubMed
-
- Bamps S, Westerling T, Pihlak A, Tafforeau L, Vandenhaute J, Mäkelä TP, Hermand D. Mcs2 and a novel CAK subunit Pmh1 associate with Skp1 in fission yeast. Biochem. Biophys. Res. Commun. 2004;325:1424–1432. - PubMed
-
- Björk GR. Biosynthesis and function of modified nucleosides. In: D. S, RajBhandary U, editors. tRNA: Structure, Biosynthesis, and Foundation. American Society for Microbiology; Washington, DC: 1995. pp. 165–205.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

