Metabolic regulation of epigenetics
- PMID: 22768835
- PMCID: PMC3392647
- DOI: 10.1016/j.cmet.2012.06.001
Metabolic regulation of epigenetics
Abstract
How cells sense and respond to environmental cues remains a central question of biological research. Recent evidence suggests that DNA transcription is regulated by chromatin organization. However, the mechanism for relaying the cytoplasmic signaling to chromatin remodeling remains incompletely understood. Although much emphasis has been put on delineating transcriptional output of growth factor/hormonal signaling pathways, accumulated evidence from yeast and mammalian systems suggest that metabolic signals also play critical roles in determining chromatin structure. Here we summarize recent progress in understanding the molecular connection between metabolism and epigenetic modifications of chromatin implicated in a variety of diseases including cancer.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

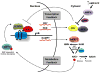
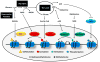

References
-
- Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, Pollock R, O’Donnell P, Grigoriadis A, Diss T, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–343. - PubMed
-
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. - PubMed
-
- Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142:943–953. - PubMed
-
- Baek SH. When signaling kinases meet histones and histone modifiers in the nucleus. Mol Cell. 2011;42:274–284. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

