A comparison of genotype-phenotype maps for RNA and proteins
- PMID: 22768948
- PMCID: PMC3328697
- DOI: 10.1016/j.bpj.2012.01.047
A comparison of genotype-phenotype maps for RNA and proteins
Abstract
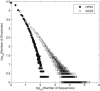
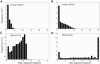
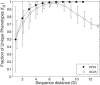
The relationship between the genotype (sequence) and the phenotype (structure) of macromolecules affects their ability to evolve new structures and functions. We here compare the genotype space organization of proteins and RNA molecules to identify differences that may affect this ability. To this end, we computationally study the genotype-phenotype relationship for short RNA and lattice proteins of a reduced monomer alphabet size, to make exhaustive analysis and direct comparison of their genotype spaces feasible. We find that many fewer protein molecules than RNA molecules fold, but they fold into many more structures than RNA. In consequence, protein phenotypes have smaller genotype networks whose member genotypes tend to be more similar than for RNA phenotypes. Neighborhoods in sequence space of a given radius around an RNA molecule contain more novel structures than for protein molecules. We compare this property to evidence from natural RNA and protein molecules, and conclude that RNA genotype space may be more conducive to the evolution of new structure phenotypes.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Wilson D.S., Szostak J.W. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. - PubMed
-
- Smith J.M. Natural selection and the concept of a protein space. Nature. 1970;225:563–564. - PubMed
-
- Lau K.F., Dill K.A. A lattice statistical mechanics model of the conformational and sequence spaces of proteins. Macromolecules. 1989;22:3986–3997.
-
- Buchler N.E.G., Goldstein R.A. Effect of alphabet size and foldability requirements on protein structure designability. Proteins. 1999;34:113–124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

