Molecular basis for the distinct divalent cation requirement in the uridylylation of the signal transduction proteins GlnJ and GlnB from Rhodospirillum rubrum
- PMID: 22769741
- PMCID: PMC3480911
- DOI: 10.1186/1471-2180-12-136
Molecular basis for the distinct divalent cation requirement in the uridylylation of the signal transduction proteins GlnJ and GlnB from Rhodospirillum rubrum
Abstract
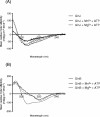
Background: PII proteins have a fundamental role in the control of nitrogen metabolism in bacteria, through interactions with different PII targets, controlled by metabolite binding and post-translational modification, uridylylation in most organisms. In the photosynthetic bacterium Rhodospirillum rubrum, the PII proteins GlnB and GlnJ were shown, in spite of their high degree of similarity, to have different requirements for post-translational uridylylation, with respect to the divalent cations, Mg(2+) and Mn(2+).
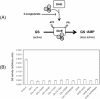
Results: Given the importance of uridylylation in the functional interactions of PII proteins, we have hypothesized that the difference in the divalent cation requirement for the uridylylation is related to efficient binding of Mg/Mn-ATP to the PII proteins. We concluded that the amino acids at positions 42 and 85 in GlnJ and GlnB (in the vicinity of the ATP binding site) influence the divalent cation requirement for uridylylation catalyzed by GlnD.
Conclusions: Efficient binding of Mg/Mn-ATP to the PII proteins is required for uridylylation by GlnD. Our results show that by simply exchanging two amino acid residues, we could modulate the divalent cation requirement in the uridylylation of GlnJ and GlnB.Considering that post-translational uridylylation of PII proteins modulates their signaling properties, a different requirement for divalent cations in the modification of GlnB and GlnJ adds an extra regulatory layer to the already intricate control of PII function.
Figures



References
-
- Zimmer DP, Soupene E, Lee HL, Wendisch VF, Khodursky AB, Peter BJ, Bender RA, Kustu S. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc Natl Acad Sci U S A. 2000;97(26):14674–14679. doi: 10.1073/pnas.97.26.14674. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

