Substrate specificity of mammalian N-terminal α-amino methyltransferase NRMT
- PMID: 22769851
- PMCID: PMC3447998
- DOI: 10.1021/bi300278f
Substrate specificity of mammalian N-terminal α-amino methyltransferase NRMT
Abstract
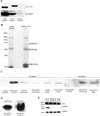
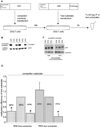
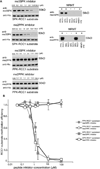
N-Terminal methylation of free α-amino groups is a post-translational modification of proteins that was first described 30 years ago but has been studied very little. In this modification, the initiating M residue is cleaved and the exposed α-amino group is mono-, di-, or trimethylated by NRMT, a recently identified N-terminal methyltransferase. Currently, all known eukaryotic α-amino-methylated proteins have a unique N-terminal motif, M-X-P-K, where X is A, P, or S. NRMT can also methylate artificial substrates in vitro in which X is G, F, Y, C, M, K, R, N, Q, or H. Methylation efficiencies of N-terminal amino acids are variable with respect to the identity of X. Here we use in vitro peptide methylation assays and substrate immunoprecipitations to show that the canonical M-X-P-K methylation motif is not the only one recognized by NRMT. We predict that N-terminal methylation is a widespread post-translational modification and that there is interplay between N-terminal acetylation and N-terminal methylation. We also use isothermal calorimetry experiments to demonstrate that NRMT can efficiently recognize and bind to its fully methylated products.
Figures

References
-
- Tsunasawa S, Stewart JW, Sherman F. Amino-terminal processing of mutant forms of yeast iso-1-cytochrome c. The specificities of methionine aminopeptidase and acetyltransferase. J Biol Chem. 1985;260(9):5382–5391. - PubMed
-
- Stock A, Clarke S, Clarke C, Stock J. N-terminal methylation of proteins: structure, function and specificity. FEBS Lett. 1987;220(1):8–14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

