U1 snRNP determines mRNA length and regulates isoform expression
- PMID: 22770214
- PMCID: PMC3412174
- DOI: 10.1016/j.cell.2012.05.029
U1 snRNP determines mRNA length and regulates isoform expression
Abstract
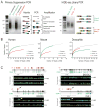
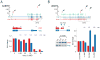
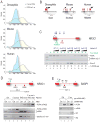
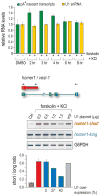
U1 snRNP (U1), in addition to its splicing role, protects pre-mRNAs from drastic premature termination by cleavage and polyadenylation (PCPA) at cryptic polyadenylation signals (PASs) in introns. Here, a high-throughput sequencing strategy of differentially expressed transcripts (HIDE-seq) mapped PCPA sites genome wide in divergent organisms. Surprisingly, whereas U1 depletion terminated most nascent gene transcripts within ~1 kb, moderate functional U1 level decreases, insufficient to inhibit splicing, dose-dependently shifted PCPA downstream and elicited mRNA 3' UTR shortening and proximal 3' exon switching characteristic of activated immune and neuronal cells, stem cells, and cancer. Activated neurons' signature mRNA shortening could be recapitulated by U1 decrease and antagonized by U1 overexpression. Importantly, we show that rapid and transient transcriptional upregulation inherent to neuronal activation physiology creates U1 shortage relative to pre-mRNAs. Additional experiments suggest cotranscriptional PCPA counteracted by U1 association with nascent transcripts, a process we term telescripting, ensuring transcriptome integrity and regulating mRNA length.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures



Comment in
-
U1 snRNA rewrites the "script".Cell. 2012 Jul 6;150(1):9-11. doi: 10.1016/j.cell.2012.06.017. Cell. 2012. PMID: 22770211 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

