The LIM protein Ajuba restricts the second heart field progenitor pool by regulating Isl1 activity
- PMID: 22771034
- PMCID: PMC3671491
- DOI: 10.1016/j.devcel.2012.06.005
The LIM protein Ajuba restricts the second heart field progenitor pool by regulating Isl1 activity
Abstract
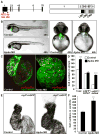
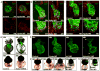
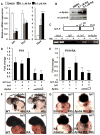
Morphogenesis of the heart requires tight control of cardiac progenitor cell specification, expansion, and differentiation. Retinoic acid (RA) signaling restricts expansion of the second heart field (SHF), serving as an important morphogen in heart development. Here, we identify the LIM domain protein Ajuba as a crucial regulator of the SHF progenitor cell specification and expansion. Ajuba-deficient zebrafish embryos show an increased pool of Isl1(+) cardiac progenitors and, subsequently, dramatically increased numbers of cardiomyocytes at the arterial and venous poles. Furthermore, we show that Ajuba binds Isl1, represses its transcriptional activity, and is also required for autorepression of Isl1 expression in an RA-dependent manner. Lack of Ajuba abrogates the RA-dependent restriction of Isl1(+) cardiac cells. We conclude that Ajuba plays a central role in regulating the SHF during heart development by linking RA signaling to the function of Isl1, a key transcription factor in cardiac progenitor cells.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
A BMP4-p38 MAPK signaling axis controls ISL1 protein stability and activity during cardiogenesis.Stem Cell Reports. 2021 Aug 10;16(8):1894-1905. doi: 10.1016/j.stemcr.2021.06.017. Epub 2021 Jul 29. Stem Cell Reports. 2021. PMID: 34329593 Free PMC article.
-
The Isl1/Ldb1 Complex Orchestrates Genome-wide Chromatin Organization to Instruct Differentiation of Multipotent Cardiac Progenitors.Cell Stem Cell. 2015 Sep 3;17(3):287-99. doi: 10.1016/j.stem.2015.08.007. Epub 2015 Aug 27. Cell Stem Cell. 2015. PMID: 26321200 Free PMC article.
-
Retinoic acid signaling restricts the size of the first heart field within the anterior lateral plate mesoderm.Dev Biol. 2021 May;473:119-129. doi: 10.1016/j.ydbio.2021.02.005. Epub 2021 Feb 16. Dev Biol. 2021. PMID: 33607112 Free PMC article.
-
Islet1-expressing cardiac progenitor cells: a comparison across species.Dev Genes Evol. 2013 Mar;223(1-2):117-29. doi: 10.1007/s00427-012-0400-1. Epub 2012 Apr 24. Dev Genes Evol. 2013. PMID: 22526874 Free PMC article. Review.
-
Mechanisms of retinoic acid signaling during cardiogenesis.Mech Dev. 2017 Feb;143:9-19. doi: 10.1016/j.mod.2016.12.002. Epub 2016 Dec 19. Mech Dev. 2017. PMID: 28007475 Review.
Cited by
-
Impaired development of left anterior heart field by ectopic retinoic acid causes transposition of the great arteries.J Am Heart Assoc. 2015 Apr 30;4(5):e001889. doi: 10.1161/JAHA.115.001889. J Am Heart Assoc. 2015. PMID: 25929268 Free PMC article.
-
Islet1 is a direct transcriptional target of the homeodomain transcription factor Shox2 and rescues the Shox2-mediated bradycardia.Basic Res Cardiol. 2013 Mar;108(2):339. doi: 10.1007/s00395-013-0339-z. Epub 2013 Mar 1. Basic Res Cardiol. 2013. PMID: 23455426 Free PMC article.
-
Bi-FoRe: an efficient bidirectional knockin strategy to generate pairwise conditional alleles with fluorescent indicators.Protein Cell. 2021 Jan;12(1):39-56. doi: 10.1007/s13238-020-00747-1. Epub 2020 Jul 17. Protein Cell. 2021. PMID: 32681448 Free PMC article.
-
Wnt9 directs zebrafish heart tube assembly via a combination of canonical and non-canonical pathway signaling.Development. 2023 Sep 15;150(18):dev201707. doi: 10.1242/dev.201707. Epub 2023 Sep 25. Development. 2023. PMID: 37680191 Free PMC article.
-
Pioneering function of Isl1 in the epigenetic control of cardiomyocyte cell fate.Cell Res. 2019 Jun;29(6):486-501. doi: 10.1038/s41422-019-0168-1. Epub 2019 Apr 25. Cell Res. 2019. PMID: 31024170 Free PMC article.
References
-
- Azpiazu N, Frasch M. tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 1993;7(7B):1325–1340. - PubMed
-
- Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118:719–729. - PubMed
-
- Brade T, Gessert S, Kühl M, Pandur P. The amphibian second heart field: Xenopus islet-1 is required for cardiovascular development. Dev Biol. 2007;311:297–310. - PubMed
-
- Brand T. Heart development: molecular insights into cardiac specification and early morphogenesis. Dev Biol. 2003;258:1–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

