Small terminase couples viral DNA binding to genome-packaging ATPase activity
- PMID: 22771211
- PMCID: PMC3563279
- DOI: 10.1016/j.str.2012.05.014
Small terminase couples viral DNA binding to genome-packaging ATPase activity
Abstract
Packaging of viral genomes into empty procapsids is powered by a large DNA-packaging motor. In most viruses, this machine is composed of a large (L) and a small (S) terminase subunit complexed with a dodecamer of portal protein. Here we describe the 1.75 Å crystal structure of the bacteriophage P22 S-terminase in a nonameric conformation. The structure presents a central channel ∼23 Å in diameter, sufficiently large to accommodate hydrated B-DNA. The last 23 residues of S-terminase are essential for binding to DNA and assembly to L-terminase. Upon binding to its own DNA, S-terminase functions as a specific activator of L-terminase ATPase activity. The DNA-dependent stimulation of ATPase activity thus rationalizes the exclusive specificity of genome-packaging motors for viral DNA in the crowd of host DNA, ensuring fidelity of packaging and avoiding wasteful ATP hydrolysis. This posits a model for DNA-dependent activation of genome-packaging motors of general interest in virology.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures


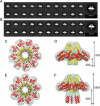

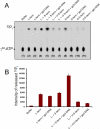
Comment in
-
Themes and variations of viral small terminase proteins.Structure. 2012 Aug 8;20(8):1291-2. doi: 10.1016/j.str.2012.07.007. Structure. 2012. PMID: 22884105 Free PMC article.
Similar articles
-
Crystallization of the nonameric small terminase subunit of bacteriophage P22.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011 Jan 1;67(Pt 1):104-10. doi: 10.1107/S174430911004697X. Epub 2010 Dec 23. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011. PMID: 21206037 Free PMC article.
-
Architecture of the Complex Formed by Large and Small Terminase Subunits from Bacteriophage P22.J Mol Biol. 2015 Oct 9;427(20):3285-3299. doi: 10.1016/j.jmb.2015.08.013. Epub 2015 Aug 21. J Mol Biol. 2015. PMID: 26301600 Free PMC article.
-
Nucleotides regulate the conformational state of the small terminase subunit from bacteriophage lambda: implications for the assembly of a viral genome-packaging motor.Biochemistry. 2005 Jul 19;44(28):9645-56. doi: 10.1021/bi050333e. Biochemistry. 2005. PMID: 16008350
-
The bacteriophage DNA packaging machine.Adv Exp Med Biol. 2012;726:489-509. doi: 10.1007/978-1-4614-0980-9_22. Adv Exp Med Biol. 2012. PMID: 22297528 Review.
-
The bacteriophage DNA packaging motor.Annu Rev Genet. 2008;42:647-81. doi: 10.1146/annurev.genet.42.110807.091545. Annu Rev Genet. 2008. PMID: 18687036 Review.
Cited by
-
Novel DNA packaging recognition in the unusual bacteriophage N15.Virology. 2015 Aug;482:260-8. doi: 10.1016/j.virol.2015.03.027. Epub 2015 May 16. Virology. 2015. PMID: 25956737 Free PMC article.
-
Enteric Chromosomal Islands: DNA Packaging Specificity and Role of λ-like Helper Phage Terminase.Viruses. 2022 Apr 15;14(4):818. doi: 10.3390/v14040818. Viruses. 2022. PMID: 35458547 Free PMC article.
-
Biophysical and structural characterization of a multifunctional viral genome packaging motor.Nucleic Acids Res. 2024 Jan 25;52(2):831-843. doi: 10.1093/nar/gkad1135. Nucleic Acids Res. 2024. PMID: 38084901 Free PMC article.
-
Large terminase conformational change induced by connector binding in bacteriophage T7.J Biol Chem. 2013 Jun 7;288(23):16998-17007. doi: 10.1074/jbc.M112.448951. Epub 2013 Apr 30. J Biol Chem. 2013. PMID: 23632014 Free PMC article.
-
Recognition of an α-helical hairpin in P22 large terminase by a synthetic antibody fragment.Acta Crystallogr D Struct Biol. 2020 Sep 1;76(Pt 9):876-888. doi: 10.1107/S2059798320009912. Epub 2020 Aug 17. Acta Crystallogr D Struct Biol. 2020. PMID: 32876063 Free PMC article.
References
-
- Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42.
-
- Ackermann HW. Bacteriophage observations and evolution. Res Microbiol. 2003;154:245–251. - PubMed
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. - PubMed
-
- Baumann RG, Black LW. Isolation and characterization of T4 bacteriophage gp17 terminase, a large subunit multimer with enhanced ATPase activity. J Biol Chem. 2003;278:4618–4627. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

