A role for intersubunit interactions in maintaining SAGA deubiquitinating module structure and activity
- PMID: 22771212
- PMCID: PMC3423971
- DOI: 10.1016/j.str.2012.05.015
A role for intersubunit interactions in maintaining SAGA deubiquitinating module structure and activity
Abstract
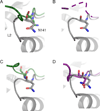
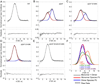
The deubiquitinating module (DUBm) of the SAGA coactivator contains the Ubp8 isopeptidase, Sgf11, Sus1, and Sgf73, which form a highly interconnected complex. Although Ubp8 contains a canonical USP catalytic domain, it is only active when in complex with the other DUBm subunits. The Sgf11 zinc finger (Sgf11-ZnF) binds near the Ubp8 active site and is essential for full activity, suggesting that the Sgf11-ZnF helps maintain the active conformation of Ubp8. We report structural and solution studies showing that deletion of the Sgf11-ZnF destabilizes incorporation of Ubp8 within the DUBm, giving rise to domain swapping with a second complex and misaligning active site residues. Activating mutations in Ubp8 that partially restore activity in the absence of the Sgf11-ZnF promote the monomeric form of the DUBm. Our data suggest an unexpected role for Sgf11 in compensating for the absence of structural features that maintain the active conformation of Ubp8.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
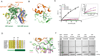



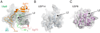
Similar articles
-
DNA binding by Sgf11 protein affects histone H2B deubiquitination by Spt-Ada-Gcn5-acetyltransferase (SAGA).J Biol Chem. 2014 Mar 28;289(13):8989-99. doi: 10.1074/jbc.M113.500868. Epub 2014 Feb 7. J Biol Chem. 2014. PMID: 24509845 Free PMC article.
-
Uncovering the role of Sgf73 in maintaining SAGA deubiquitinating module structure and activity.J Mol Biol. 2015 Apr 24;427(8):1765-78. doi: 10.1016/j.jmb.2014.12.004. Epub 2014 Dec 17. J Mol Biol. 2015. PMID: 25526805 Free PMC article.
-
Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module.Cell. 2010 May 14;141(4):606-17. doi: 10.1016/j.cell.2010.04.026. Epub 2010 Apr 29. Cell. 2010. PMID: 20434206 Free PMC article.
-
SAGA-CORE subunit Spt7 is required for correct Ubp8 localization, chromatin association and deubiquitinase activity.Epigenetics Chromatin. 2020 Oct 28;13(1):46. doi: 10.1186/s13072-020-00367-3. Epigenetics Chromatin. 2020. PMID: 33115507 Free PMC article.
-
The promiscuity of the SAGA complex subunits: Multifunctional or moonlighting proteins?Biochim Biophys Acta Gene Regul Mech. 2021 Feb;1864(2):194607. doi: 10.1016/j.bbagrm.2020.194607. Epub 2020 Jul 23. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 32712338 Review.
Cited by
-
Whole-Genome Sequencing of Suppressor DNA Mixtures Identifies Pathways That Compensate for Chromosome Segregation Defects in Schizosaccharomyces pombe.G3 (Bethesda). 2018 Mar 2;8(3):1031-1038. doi: 10.1534/g3.118.200048. G3 (Bethesda). 2018. PMID: 29352077 Free PMC article.
-
Transient interdomain interactions in free USP14 shape its conformational ensemble.Protein Sci. 2024 May;33(5):e4975. doi: 10.1002/pro.4975. Protein Sci. 2024. PMID: 38588275 Free PMC article.
-
Sharing the SAGA.Trends Biochem Sci. 2017 Nov;42(11):850-861. doi: 10.1016/j.tibs.2017.09.001. Epub 2017 Sep 27. Trends Biochem Sci. 2017. PMID: 28964624 Free PMC article. Review.
-
The histone H4 basic patch regulates SAGA-mediated H2B deubiquitination and histone acetylation.J Biol Chem. 2020 May 8;295(19):6561-6569. doi: 10.1074/jbc.RA120.013196. Epub 2020 Apr 3. J Biol Chem. 2020. PMID: 32245891 Free PMC article.
-
Sus1p facilitates pre-initiation complex formation at the SAGA-regulated genes independently of histone H2B de-ubiquitylation.J Mol Biol. 2014 Aug 12;426(16):2928-2941. doi: 10.1016/j.jmb.2014.05.028. Epub 2014 Jun 6. J Mol Biol. 2014. PMID: 24911582 Free PMC article.
References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. Sect. D: Biol. Crystallogr. 2002;58:1948–1954. - PubMed
-
- Avvakumov GV, Walker JR, Xue S, Finerty PJ, Mackenzie F, Newman EM, Dhe-Paganon S. Amino-terminal dimerization, NRDP1-rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8) J. Biol. Chem. 2006;281:38061–38070. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

