Design, synthesis, biochemical studies, cellular characterization, and structure-based computational studies of small molecules targeting the urokinase receptor
- PMID: 22771232
- PMCID: PMC3437670
- DOI: 10.1016/j.bmc.2012.06.002
Design, synthesis, biochemical studies, cellular characterization, and structure-based computational studies of small molecules targeting the urokinase receptor
Abstract
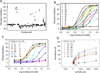
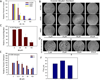
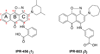
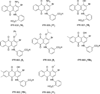
The urokinase receptor (uPAR) serves as a docking site to the serine protease urokinase-type plasminogen activator (uPA) to promote extracellular matrix (ECM) degradation and tumor invasion and metastasis. Previously, we had reported a small molecule inhibitor of the uPAR·uPA interaction that emerged from structure-based virtual screening. Here, we measure the affinity of a large number of derivatives from commercial sources. Synthesis of additional compounds was carried out to probe the role of various groups on the parent compound. Extensive structure-based computational studies suggested a binding mode for these compounds that led to a structure-activity relationship study. Cellular studies in non-small cell lung cancer (NSCLC) cell lines that include A549, H460 and H1299 showed that compounds blocked invasion, migration and adhesion. The effects on invasion of active compounds were consistent with their inhibition of uPA and MMP proteolytic activity. These compounds showed weak cytotoxicity consistent with the confined role of uPAR to metastasis.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures







References
-
- Werle B, Kotzsch M, Lah TT, Kos J, Gabrijelcic-Geiger D, Spiess E, Schirren J, Ebert W, Fiehn W, Luther T, Magdolen V, Schmitt M, Harbeck N. Anticancer research. 2004;24:4147. - PubMed
-
- Andreasen PA, Kjoller L, Christensen L, Duffy MJ. Int J Cancer. 1997;72:1. - PubMed
-
- Wei Y, Tang CH, Kim Y, Robillard L, Zhang F, Kugler MC, Chapman HA. J Biol Chem. 2007;282:3929. - PubMed
-
- Simon DI, Wei Y, Zhang L, Rao NK, Xu H, Chen ZP, Liu QM, Rosenberg S, Chapman HA. J. Biol. Chem. 2000;275:10228. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Miscellaneous

