A peroxisomally localized acyl-activating enzyme is required for volatile benzenoid formation in a Petuniaxhybrida cv. 'Mitchell Diploid' flower
- PMID: 22771854
- PMCID: PMC3428004
- DOI: 10.1093/jxb/ers153
A peroxisomally localized acyl-activating enzyme is required for volatile benzenoid formation in a Petuniaxhybrida cv. 'Mitchell Diploid' flower
Abstract
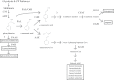
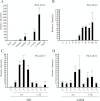
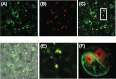
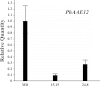
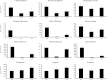
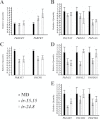
Floral volatile benzenoid/phenylpropanoid (FVBP) biosynthesis is a complex and coordinate cellular process executed by petal limb cells of a Petunia×hybrida cv. 'Mitchell Diploid' (MD) plant. In MD flowers, the majority of benzenoid volatile compounds are derived from a core phenylpropanoid pathway intermediate by a coenzyme A (CoA) dependent, β-oxidative scheme. Metabolic flux analysis, reverse genetics, and biochemical characterizations of key enzymes in this pathway have supported this putative concept. However, the theoretical first enzymatic reaction, which leads to the production of cinnamoyl-CoA, has only been physically demonstrated in a select number of bacteria like Streptomyces maritimus through mutagenesis and recombinant protein production. A transcript has been cloned and characterized from MD flowers that shares high homology with an Arabidopsis thaliana transcript ACYL-ACTIVATING ENZYME11 (AtAAE11) and the S. maritimus ACYL-COA:LIGASE (SmEncH). In MD, the PhAAE transcript accumulates in a very similar manner as bona fide FVBP network genes, i.e. high levels in an open flower petal and ethylene regulated. In planta, PhAAE is localized to the peroxisome. Upon reduction of PhAAE transcript through a stable RNAi approach, transgenic flowers emitted a reduced level of all benzenoid volatile compounds. Together, the data suggest that PhAAE may be responsible for the activation of t-cinnamic acid, which would be required for floral volatile benzenoid production in MD.
Figures

References
-
- Anterola AM, Jeon JH, Davin LB, Lewis NG. Transcriptional control of monolignol biosynthesis in Pinus taeda: factors affecting monolignol ratios and carbon allocation in phenylpropanoid metabolism. Journal of Biological Chemistry. 2002;277:18272–18280. - PubMed
-
- Beuerle T, Pichersky E. Purification and characterization of benzoate:coenzyme A ligase from Clarkia breweri . Archives of Biochemistry and Biochemistry. 2002;400:258–264. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

