Transcription regulation by distal enhancers: who's in the loop?
- PMID: 22771987
- PMCID: PMC3654767
- DOI: 10.4161/trns.20720
Transcription regulation by distal enhancers: who's in the loop?
Abstract
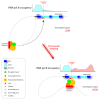
Genome-wide chromatin profiling efforts have shown that enhancers are often located at large distances from gene promoters within the noncoding genome. Whereas enhancers can stimulate transcription initiation by communicating with promoters via chromatin looping mechanisms, we propose that enhancers may also stimulate transcription elongation by physical interactions with intronic elements. We review here recent findings derived from the study of the hematopoietic system.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
