The BAF complex and HIV latency
- PMID: 22771990
- PMCID: PMC3654765
- DOI: 10.4161/trns.20541
The BAF complex and HIV latency
Abstract
The persistence of a reservoir of transcriptionally competent but latent virus in the presence of antiviral regimens presents the main impediment to a curative therapy against HIV. Therefore it is critical to understand the molecular mechanisms, which lead to the establishment and maintenance of HIV latency, and which contribute to the reversal of this process and mediate HIV transcriptional activation in response to T cell activation signals. Here I discuss features of the nucleosomal landscape of the HIV promoter or 5'LTR in controlling HIV transcription. I emphasize on the emerging understanding of the role of the ATP dependent SWI/SNF chromatin remodelling complexes in modulating the chromatin architecture at the HIV LTR and how this leads to a tight regulation of LTR transcription.
Figures
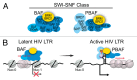
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
