Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis
- PMID: 22772370
- PMCID: PMC3702264
- DOI: 10.1038/ng.2344
Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis
Abstract
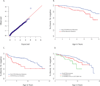
Exome sequencing has become a powerful and effective strategy for the discovery of genes underlying Mendelian disorders. However, use of exome sequencing to identify variants associated with complex traits has been more challenging, partly because the sample sizes needed for adequate power may be very large. One strategy to increase efficiency is to sequence individuals who are at both ends of a phenotype distribution (those with extreme phenotypes). Because the frequency of alleles that contribute to the trait are enriched in one or both phenotype extremes, a modest sample size can potentially be used to identify novel candidate genes and/or alleles. As part of the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP), we used an extreme phenotype study design to discover that variants in DCTN4, encoding a dynactin protein, are associated with time to first P. aeruginosa airway infection, chronic P. aeruginosa infection and mucoid P. aeruginosa in individuals with cystic fibrosis.
Figures
References
-
- Bamshad MJ, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nature reviews. Genetics. 2011;12:745–755. - PubMed
-
- Gibson RL, Burns JL, Ramsey BW. Pathophysiology and management of pulmonary infections in cystic fibrosis. American journal of respiratory and critical care medicine. 2003;168:918–951. - PubMed
-
- Emerson J, Rosenfeld M, McNamara S, Ramsey B, Gibson RL. Pseudomonas aeruginosa and other predictors of mortality and morbidity in young children with cystic fibrosis. Pediatric pulmonology. 2002;34:91–100. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K99 HG004316/HG/NHGRI NIH HHS/United States
- RC2 HL102923/HL/NHLBI NIH HHS/United States
- RC2 HL-102926/HL/NHLBI NIH HHS/United States
- G0701075/MRC_/Medical Research Council/United Kingdom
- P30 DK089507/DK/NIDDK NIH HHS/United States
- RC2 HL-102923/HL/NHLBI NIH HHS/United States
- 5R01 HG004316/HG/NHGRI NIH HHS/United States
- UL1 TR000439/TR/NCATS NIH HHS/United States
- 090532/WT_/Wellcome Trust/United Kingdom
- RC2 HL-102925/HL/NHLBI NIH HHS/United States
- RC2 HL103010/HL/NHLBI NIH HHS/United States
- RC2 HL102926/HL/NHLBI NIH HHS/United States
- R00 HG004316/HG/NHGRI NIH HHS/United States
- RC2 HL-102924/HL/NHLBI NIH HHS/United States
- RC2 HL102924/HL/NHLBI NIH HHS/United States
- RC2 HL102925/HL/NHLBI NIH HHS/United States
- R01 HL068890/HL/NHLBI NIH HHS/United States
- RC2 HL-103010/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

