Identification of new microRNA-regulated genes by conserved targeting in plant species
- PMID: 22772987
- PMCID: PMC3467045
- DOI: 10.1093/nar/gks625
Identification of new microRNA-regulated genes by conserved targeting in plant species
Abstract
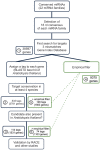
MicroRNAs (miRNAs) are major regulators of gene expression in multicellular organisms. They recognize their targets by sequence complementarity and guide them to cleavage or translational arrest. It is generally accepted that plant miRNAs have extensive complementarity to their targets and their prediction usually relies on the use of empirical parameters deduced from known miRNA-target interactions. Here, we developed a strategy to identify miRNA targets which is mainly based on the conservation of the potential regulation in different species. We applied the approach to expressed sequence tags datasets from angiosperms. Using this strategy, we predicted many new interactions and experimentally validated previously unknown miRNA targets in Arabidopsis thaliana. Newly identified targets that are broadly conserved include auxin regulators, transcription factors and transporters. Some of them might participate in the same pathways as the targets known before, suggesting that some miRNAs might control different aspects of a biological process. Furthermore, this approach can be used to identify targets present in a specific group of species, and, as a proof of principle, we analyzed Solanaceae-specific targets. The presented strategy can be used alone or in combination with other approaches to find miRNA targets in plants.
Figures





References
-
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. - PubMed
-
- Mateos JL, Bologna NG, Chorostecki U, Palatnik JF. Identification of microRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Curr. Biol. 2010;20:49–54. - PubMed
-
- Song L, Axtell MJ, Fedoroff NV. RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr. Biol. 2010;20:37–41. - PubMed
-
- Werner S, Wollmann H, Schneeberger K, Weigel D. Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr. Biol. 2010;20:42–48. - PubMed

