Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex
- PMID: 22773744
- PMCID: PMC3426135
- DOI: 10.1105/tpc.112.099796
Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex
Abstract
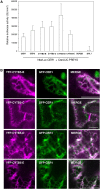
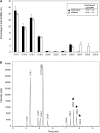
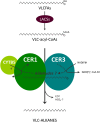
In land plants, very-long-chain (VLC) alkanes are major components of cuticular waxes that cover aerial organs, mainly acting as a waterproof barrier to prevent nonstomatal water loss. Although thoroughly investigated, plant alkane synthesis remains largely undiscovered. The Arabidopsis thaliana ECERIFERUM1 (CER1) protein has been recognized as an essential element of wax alkane synthesis; nevertheless, its function remains elusive. In this study, a screen for CER1 physical interaction partners was performed. The screen revealed that CER1 interacts with the wax-associated protein ECERIFERUM3 (CER3) and endoplasmic reticulum-localized cytochrome b5 isoforms (CYTB5s). The functional relevance of these interactions was assayed through an iterative approach using yeast as a heterologous expression system. In a yeast strain manipulated to produce VLC acyl-CoAs, a strict CER1 and CER3 coexpression resulted in VLC alkane synthesis. The additional presence of CYTB5s was found to enhance CER1/CER3 alkane production. Site-directed mutagenesis showed that CER1 His clusters are essential for alkane synthesis, whereas those of CER3 are not, suggesting that CYTB5s are specific CER1 cofactors. Collectively, our study reports the identification of plant alkane synthesis enzymatic components and supports a new model for alkane production in which CER1 interacts with both CER3 and CYTB5 to catalyze the redox-dependent synthesis of VLC alkanes from VLC acyl-CoAs.
Figures



References
-
- Bourdenx B., Bernard A., Domergue F., Pascal S., Léger A., Roby D., Pervent M., Vile D., Haslam R.P., Napier J.A., Lessire R., Joubès J. (2011). Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 156: 29–45 - PMC - PubMed
-
- Buschhaus C., Jetter R. (2011). Composition differences between epicuticular and intracuticular wax substructures: How do plants seal their epidermal surfaces? J. Exp. Bot. 62: 841–853 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

