Genome-wide identification of regulatory DNA elements and protein-binding footprints using signatures of open chromatin in Arabidopsis
- PMID: 22773751
- PMCID: PMC3426110
- DOI: 10.1105/tpc.112.098061
Genome-wide identification of regulatory DNA elements and protein-binding footprints using signatures of open chromatin in Arabidopsis
Abstract
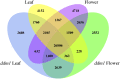
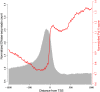
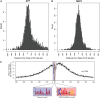
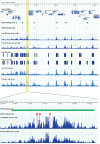
Gene expression and regulation in eukaryotes is controlled by orchestrated binding of regulatory proteins, including both activators and repressors, to promoters and other cis-regulatory DNA elements. An increasing number of plant genomes have been sequenced; however, a similar effort to the ENCODE project, which aimed to identify all functional elements in the human genome, has yet to be initiated in plants. Here we report genome-wide high-resolution mapping of DNase I hypersensitive (DH) sites in the model plant Arabidopsis thaliana. We identified 38,290 and 41,193 DH sites in leaf and flower tissues, respectively. The DH sites were depleted of bulk nucleosomes and were tightly associated with RNA polymerase II binding sites. Approximately 90% of the binding sites of two well-characterized MADS domain transcription factors, APETALA1 and SEPALLATA3, were covered by the DH sites. We demonstrate that protein binding footprints within a specific genomic region can be revealed using the DH site data sets in combination with known or putative protein binding motifs and gene expression data sets. Thus, genome-wide DH site mapping will be an important tool for systematic identification of all cis-regulatory DNA elements in plants.
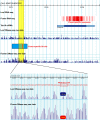
Figures




References
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
-
- Ausubel F.M. (2002). Summaries of National Science Foundation-sponsored Arabidopsis 2010 projects and National Science Foundation-sponsored plant genome projects that are generating Arabidopsis resources for the community. Plant Physiol. 129: 394–437
-
- Bailey T.L., Elkan C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2: 28–36 - PubMed
-
- Birnbaum K., Shasha D.E., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N. (2003). A gene expression map of the Arabidopsis root. Science 302: 1956–1960 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

