TolC-dependent secretion of an ankyrin repeat-containing protein of Rickettsia typhi
- PMID: 22773786
- PMCID: PMC3430354
- DOI: 10.1128/JB.00793-12
TolC-dependent secretion of an ankyrin repeat-containing protein of Rickettsia typhi
Abstract
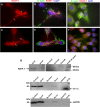
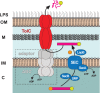
Rickettsia typhi, the causative agent of murine (endemic) typhus, is an obligate intracellular pathogen with a life cycle involving both vertebrate and invertebrate hosts. In this study, we characterized a gene (RT0218) encoding a C-terminal ankyrin repeat domain-containing protein, named Rickettsia ankyrin repeat protein 1 (RARP-1), and identified it as a secreted effector protein of R. typhi. RT0218 showed differential transcript abundance at various phases of R. typhi intracellular growth. RARP-1 was secreted by R. typhi into the host cytoplasm during in vitro infection of mammalian cells. Transcriptional analysis revealed that RT0218 was cotranscribed with adjacent genes RT0217 (hypothetical protein) and RT0216 (TolC) as a single polycistronic mRNA. Given one of its functions as a facilitator of extracellular protein secretion in some Gram-negative bacterial pathogens, we tested the possible role of TolC in the secretion of RARP-1. Using Escherichia coli C600 and an isogenic tolC insertion mutant as surrogate hosts, our data demonstrate that RARP-1 is secreted in a TolC-dependent manner. Deletion of either the N-terminal signal peptide or the C-terminal ankyrin repeats abolished RARP-1 secretion by wild-type E. coli. Importantly, expression of R. typhi tolC in the E. coli tolC mutant restored the secretion of RARP-1, suggesting that TolC has a role in RARP-1 translocation across the outer membrane. This work implies that the TolC component of the putative type 1 secretion system of R. typhi is involved in the secretion process of RARP-1.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

