Contribution of the C-terminal regions of promyelocytic leukemia protein (PML) isoforms II and V to PML nuclear body formation
- PMID: 22773875
- PMCID: PMC3436317
- DOI: 10.1074/jbc.M112.374769
Contribution of the C-terminal regions of promyelocytic leukemia protein (PML) isoforms II and V to PML nuclear body formation
Abstract
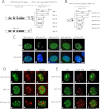
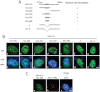
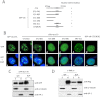
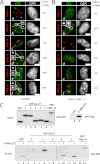
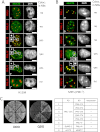
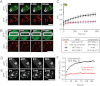
Promyelocytic leukemia protein (PML) nuclear bodies are dynamic and heterogeneous nuclear protein complexes implicated in various important functions, most notably tumor suppression. PML is the structural component of PML nuclear bodies and has several nuclear splice isoforms that share a common N-terminal region but differ in their C termini. Previous studies have suggested that the coiled-coil motif within the N-terminal region is sufficient for PML nuclear body formation by mediating homo/multi-dimerization of PML molecules. However, it has not been investigated whether any of the C-terminal variants of PML may contribute to PML body assembly. Here we report that the unique C-terminal domains of PML-II and PML-V can target to PML-NBs independent of their N-terminal region. Strikingly, both domains can form nuclear bodies in the absence of endogenous PML. The C-terminal domain of PML-II interacts transiently with unknown binding sites at PML nuclear bodies, whereas the C-terminal domain of PML-V exhibits hyperstable binding to PML bodies via homo-dimerization. This strong interaction is mediated by a putative α-helix in the C terminus of PML-V. Moreover, nuclear bodies assembled from the C-terminal domain of PML-V also recruit additional PML body components, including Daxx and Sp100. These observations establish the C-terminal domain of PML-V as an additional important contributor to the assembly mechanism(s) of PML bodies.
Figures




References
-
- Everett R. D., Lomonte P., Sternsdorf T., van Driel R., Orr A. (1999) Cell cycle regulation of PML modification and ND10 composition. J. Cell Sci. 112, 4581–4588 - PubMed
-
- Salomoni P., Pandolfi P. P. (2002) The role of PML in tumor suppression. Cell 108, 165–170 - PubMed
-
- Zhong S., Salomoni P., Pandolfi P. P. (2000) The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2, E85–E90 - PubMed
-
- Regad T., Chelbi-Alix M. K. (2001) Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene 20, 7274–7286 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

