Phylogenomics meets neuroscience: how many times might complex brains have evolved?
- PMID: 22776469
- PMCID: PMC4024470
- DOI: 10.1556/ABiol.63.2012.Suppl.2.1
Phylogenomics meets neuroscience: how many times might complex brains have evolved?
Abstract
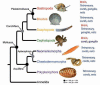
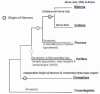
The origin of complex centralized brains is one of the major evolutionary transitions in the history of animals. Monophyly (i.e. presence of a centralized nervous system in urbilateria) vs polyphyly (i.e. multiple origins by parallel centralization of nervous systems within several lineages) are two historically conflicting scenarios to explain such transitions. However, recent phylogenomic and cladistic analysis suggests that complex brains may have independently evolved at least 9 times within different animal lineages. Indeed, even within the phylum Mollusca cephalization might have occurred at least 5 times. Emerging molecular data further suggest that at the genomic level such transitions might have been achieved by changes in expression of just a few transcriptional factors - not surprising since such events might happen multiple times over 700 million years of animal evolution. Both cladistic and genomic analyses also imply that neurons themselves evolved more than once. Ancestral polarized secretory cells were likely involved in coordination of ciliated locomotion in early animals, and these cells can be considered as evolutionary precursors of neurons within different lineages. Under this scenario, the origins of neurons can be linked to adaptations to stress/injury factors in the form of integrated regeneration-type cellular response with secretory signaling peptides as early neurotransmitters. To further reconstruct the parallel evolution of nervous systems genomic approaches are essential to probe enigmatic neurons of basal metazoans, selected lophotrochozoans (e.g. phoronids, brachiopods) and deuterostomes.
Figures




References
-
- Arendt D, Nubler-Jung K. Inversion of dorsoventral axis? Nature. 1994;371:26. - PubMed
-
- Bourlat SJ, Juliusdottir T, Lowe CJ, Freeman R, Aronowicz J, Kirschner M, lander ES, Thorndyke M, Nakano H, Kohn AB, Heyland A, Moroz LL, Copley RR, Telford MJ. Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature. 2006;444:85–88. - PubMed
-
- Bourlat SJ, Nielsen C, Lockyer AE, Littlewood DT, Telford MJ. Xenoturbella is a deuterostome that eats molluscs. Nature. 2003;424:925–928. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

