Gene duplication, tissue-specific gene expression and sexual conflict in stalk-eyed flies (Diopsidae)
- PMID: 22777023
- PMCID: PMC3391427
- DOI: 10.1098/rstb.2011.0287
Gene duplication, tissue-specific gene expression and sexual conflict in stalk-eyed flies (Diopsidae)
Abstract
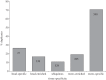
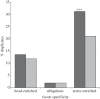
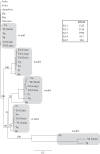
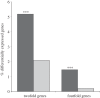
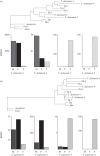
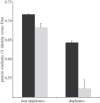
Gene duplication provides an essential source of novel genetic material to facilitate rapid morphological evolution. Traits involved in reproduction and sexual dimorphism represent some of the fastest evolving traits in nature, and gene duplication is intricately involved in the origin and evolution of these traits. Here, we review genomic research on stalk-eyed flies (Diopsidae) that has been used to examine the extent of gene duplication and its role in the genetic architecture of sexual dimorphism. Stalk-eyed flies are remarkable because of the elongation of the head into long stalks, with the eyes and antenna laterally displaced at the ends of these stalks. Many species are strongly sexually dimorphic for eyespan, and these flies have become a model system for studying sexual selection. Using both expressed sequence tag and next-generation sequencing, we have established an extensive database of gene expression in the developing eye-antennal imaginal disc, the adult head and testes. Duplicated genes exhibit narrower expression patterns than non-duplicated genes, and the testes, in particular, provide an abundant source of gene duplication. Within somatic tissue, duplicated genes are more likely to be differentially expressed between the sexes, suggesting gene duplication may provide a mechanism for resolving sexual conflict.
Figures


References
-
- Emlen D. J., Corley Lavine L., Ewen-Campen B. 2007. On the origin and evolutionary diversification of beetle horns. Proc. Natl Acad. Sci. USA 104, 8661–8668 10.1073/pnas.0701209104 (doi:10.1073/pnas.0701209104) - DOI - PMC - PubMed
-
- Hegyi G., Török J., Tóth L., Garamszegi L. Z., Rosivall B. 2006. Rapid temporal change in the expression and age-related information content of a sexually selected trait. J. Evol. Biol. 19, 228–238 10.1111/j.1420-9101.2005.00970.x (doi:10.1111/j.1420-9101.2005.00970.x) - DOI - PubMed
-
- Meyer A. 1997. The evolution of sexually selected traits in male swordtail fishes (Xiphophorus: Poeciliidae). Heredity 79, 329–337 10.1038/hdy.1997.161 (doi:10.1038/hdy.1997.161) - DOI
-
- Omland K. E., Lanyon S. M. 2000. Reconstructing plumage evolution in orioles (Icterus): repeated convergence and reversal in patterns. Evolution 54, 2119–2133 10.1111/j.0014-3820.2000.tb01254.x (doi:10.1111/j.0014-3820.2000.tb01254.x) - DOI - PubMed
-
- van Doorn G. S. 2009. Intralocus sexual conflict. Ann. NY Acad. Sci. 1168, 52–71 10.1111/j.1749-6632.2009.04573.x (doi:10.1111/j.1749-6632.2009.04573.x) - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
