Chemical and biochemical approaches in the study of histone methylation and demethylation
- PMID: 22777714
- PMCID: PMC4129395
- DOI: 10.1002/mrr.20228
Chemical and biochemical approaches in the study of histone methylation and demethylation
Abstract
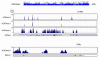
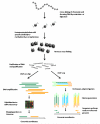
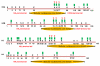
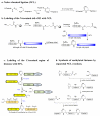
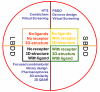
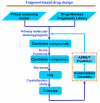
Histone methylation represents one of the most critical epigenetic events in DNA function regulation in eukaryotic organisms. Classic molecular biology and genetics tools provide significant knowledge about mechanisms and physiological roles of histone methyltransferases and demethylases in various cellular processes. In addition to this stream line, development and application of chemistry and chemistry-related techniques are increasingly involved in biological study, and offer information otherwise difficult to obtain by standard molecular biology methods. Herein, we review recent achievements and progress in developing and applying chemical and biochemical approaches in the study of histone methylation, including chromatin immunoprecipitation, chemical ligation, mass spectrometry, biochemical methylation and demethylation assays, and inhibitor development. These technological advances allow histone methylation to be studied from genome-wide level to molecular and atomic levels. With ChIP technology, information can be obtained about precise mapping of histone methylation patterns at specific promoters, genes, or other genomic regions. MS is particularly useful in detecting and analyzing methylation marks in histone and nonhistone protein substrates. Chemical approaches that permit site-specific incorporation of methyl groups into histone proteins greatly facilitate the investigation of biological impacts of methylation at individual modification sites. Discovery and design of selective organic inhibitors of histone methyltransferases and demethylases provide chemical probes to interrogate methylation-mediated cellular pathways. Overall, these chemistry-related technological advances have greatly improved our understanding of the biological functions of histone methylation in normal physiology and diseased states, and also are of great potential to translate basic epigenetics research into diagnostic and therapeutic applications in the clinic.
© 2010 Wiley Periodicals, Inc.
Figures



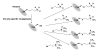



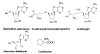



Similar articles
-
[Advances in histone methyltransferases and histone demethylases].Ai Zheng. 2008 Oct;27(10):1018-25. Ai Zheng. 2008. PMID: 18851779 Review. Chinese.
-
The emerging therapeutic potential of histone methyltransferase and demethylase inhibitors.ChemMedChem. 2009 Oct;4(10):1568-82. doi: 10.1002/cmdc.200900301. ChemMedChem. 2009. PMID: 19739196 Review.
-
Epigenetic small molecule modulators of histone and DNA methylation.Curr Opin Chem Biol. 2018 Aug;45:73-85. doi: 10.1016/j.cbpa.2018.03.003. Epub 2018 Mar 24. Curr Opin Chem Biol. 2018. PMID: 29579619 Review.
-
Assaying epigenome functions of PRMTs and their substrates.Methods. 2020 Mar 15;175:53-65. doi: 10.1016/j.ymeth.2019.09.014. Epub 2019 Sep 19. Methods. 2020. PMID: 31542509 Review.
-
Small molecule epigenetic inhibitors targeted to histone lysine methyltransferases and demethylases.Q Rev Biophys. 2013 Nov;46(4):349-73. doi: 10.1017/S0033583513000085. Epub 2013 Sep 2. Q Rev Biophys. 2013. PMID: 23991894 Free PMC article. Review.
Cited by
-
SAM/SAH Analogs as Versatile Tools for SAM-Dependent Methyltransferases.ACS Chem Biol. 2016 Mar 18;11(3):583-97. doi: 10.1021/acschembio.5b00812. Epub 2015 Nov 16. ACS Chem Biol. 2016. PMID: 26540123 Free PMC article. Review.
-
C-demethylation and 1, 2-amino shift in (E)-2-(1-(3-aminophenyl) ethylidene)hydrazinecarboxamide to (E)-2-(2-aminobenzylidene)hydrazinecarboxamide and their applications.Sci Rep. 2020 Dec 14;10(1):21913. doi: 10.1038/s41598-020-79027-1. Sci Rep. 2020. PMID: 33318572 Free PMC article.
-
DPY30 knockdown suppresses colorectal carcinoma progression via inducing Raf1/MST2-mediated apoptosis.Heliyon. 2024 Jan 20;10(3):e24807. doi: 10.1016/j.heliyon.2024.e24807. eCollection 2024 Feb 15. Heliyon. 2024. PMID: 38314299 Free PMC article.
-
The Third Intron of the Interferon Regulatory Factor-8 Is an Initiator of Repressed Chromatin Restricting Its Expression in Non-Immune Cells.PLoS One. 2016 Jun 3;11(6):e0156812. doi: 10.1371/journal.pone.0156812. eCollection 2016. PLoS One. 2016. PMID: 27257682 Free PMC article.
-
Fluorimetric and CD Recognition between Various ds-DNA/RNA Depends on a Cyanine Connectivity in Cyanine-guanidiniocarbonyl-pyrrole Conjugate.Molecules. 2020 Sep 29;25(19):4470. doi: 10.3390/molecules25194470. Molecules. 2020. PMID: 33003366 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

