FimA, FimF, and FimH are necessary for assembly of type 1 fimbriae on Salmonella enterica serovar Typhimurium
- PMID: 22778099
- PMCID: PMC3418740
- DOI: 10.1128/IAI.00331-12
FimA, FimF, and FimH are necessary for assembly of type 1 fimbriae on Salmonella enterica serovar Typhimurium
Abstract
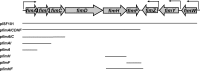
Salmonella enterica serovar Typhimurium is a Gram-negative member of the family Enterobacteriaceae and is a common cause of bacterial food poisoning in humans. The fimbrial appendages are found on the surface of many enteric bacteria and enable the bacteria to bind to eukaryotic cells. S. Typhimurium type 1 fimbriae are characterized by mannose-sensitive hemagglutination and are assembled via the chaperone/usher pathway. S. Typhimurium type 1 fimbrial proteins are encoded by the fim gene cluster (fimAICDHFZYW), with fimAICDHF expressed as a single transcriptional unit. The structural components of the fimbriae are FimA (major subunit), FimI, FimH (adhesin), and FimF (adaptor). In order to determine which components are required for fimbrial formation in S. Typhimurium, mutations in fimA, fimI, fimH, and fimF were constructed and examined for their ability to produce surface-assembled fimbriae. S. Typhimurium SL1344ΔfimA, -ΔfimH, and -ΔfimF mutants were unable to assemble fimbriae, indicating that these genes are necessary for fimbrial production in S. Typhimurium. However, SL1344ΔfimI was able to assemble fimbriae. In Escherichia coli type 1 and Pap fimbriae, at least two adaptors are expressed in addition to the adhesins. However, E. coli type 1 and Pap fimbriae have been reported to be able to assemble fimbriae in the absence of these proteins. These results suggest differences between the S. Typhimurium type 1 fimbrial system and the E. coli type 1 and Pap fimbrial systems.
Figures



References
-
- Adegbola RA, Old DC. 1987. Antigenic relationships among type-1 fimbriae of Enterobacteriaceae revealed by immuno-electronmicroscopy. J. Med. Microbiol. 24:21–28 - PubMed
-
- Blanco M, et al. 1997. Detection of pap, sfa and afa adhesin-encoding operons in uropathogenic Escherichia coli strains: relationship with expression of adhesins and production of toxins. Res. Microbiol. 148:745–755 - PubMed
-
- Boddicker JD, Ledeboer NA, Jagnow J, Jones BD, Clegg S. 2002. Differential binding to and biofilm formation on, HEp-2 cells by Salmonella enterica serovar Typhimurium is dependent upon allelic variation in the fimH gene of the fim gene cluster. Mol. Microbiol. 45:1255–1265 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

