The ubiquitin-specific protease 12 (USP12) is a negative regulator of notch signaling acting on notch receptor trafficking toward degradation
- PMID: 22778262
- PMCID: PMC3436160
- DOI: 10.1074/jbc.M112.366807
The ubiquitin-specific protease 12 (USP12) is a negative regulator of notch signaling acting on notch receptor trafficking toward degradation
Abstract
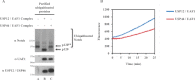
Notch signaling is critical for development and adult tissue physiology, controlling cell fate in a context-dependent manner. Upon ligand binding, the transmembrane Notch receptor undergoes two ordered proteolytic cleavages releasing Notch intracellular domain, which regulates the transcription of Notch target genes. The strength of Notch signaling is of crucial importance and depends notably on the quantity of Notch receptor at the cell surface. Using an shRNA library screen monitoring Notch trafficking and degradation in the absence of ligand, we identified mammalian USP12 and its Drosophila melanogaster homolog as novel negative regulators of Notch signaling. USP12 silencing specifically interrupts Notch trafficking to the lysosomes and, as a consequence, leads to an increased amount of receptor at the cell surface and to a higher Notch activity. At the biochemical level, USP12 with its activator UAF1 deubiquitinate the nonactivated form of Notch in cell culture and in vitro. These results characterize a new level of conserved regulation of Notch signaling by the ubiquitin system.
Figures
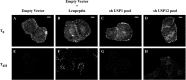

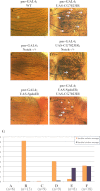
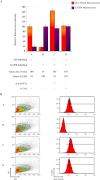
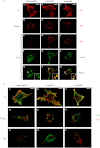


References
-
- High F. A., Epstein J. A. (2008) The multifaceted role of Notch in cardiac development and disease. Nat. Rev. Genet. 9, 49–61 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

