Up-regulation of the mitotic checkpoint component Mad1 causes chromosomal instability and resistance to microtubule poisons
- PMID: 22778409
- PMCID: PMC3421180
- DOI: 10.1073/pnas.1201911109
Up-regulation of the mitotic checkpoint component Mad1 causes chromosomal instability and resistance to microtubule poisons
Abstract
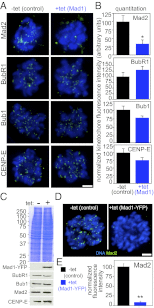
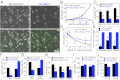
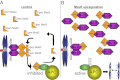
The mitotic checkpoint is the major cell cycle checkpoint acting during mitosis to prevent aneuploidy and chromosomal instability, which are hallmarks of tumor cells. Reduced expression of the mitotic checkpoint component Mad1 causes aneuploidy and promotes tumors in mice [Iwanaga Y, et al. (2007) Cancer Res 67:160-166]. However, the prevalence and consequences of Mad1 overexpression are currently unclear. Here we show that Mad1 is frequently overexpressed in human cancers and that Mad1 up-regulation is a marker of poor prognosis. Overexpression of Mad1 causes aneuploidy and chromosomal instability through weakening mitotic checkpoint signaling caused by mislocalization of the Mad1 binding partner Mad2. Cells overexpressing Mad1 are resistant to microtubule poisons, including currently used chemotherapeutic agents. These results suggest that levels of Mad1 must be tightly regulated to prevent aneuploidy and transformation and that Mad1 up-regulation may promote tumors and cause resistance to current therapies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
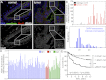
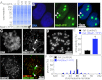
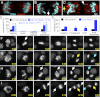
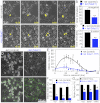


References
-
- Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol. 2006;18:658–667. - PubMed
-
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. - PubMed
-
- Rieder CL. Mitosis in vertebrates: The G2/M and M/A transitions and their associated checkpoints. Chromosome Res. 2011;19:291–306. - PubMed
-
- Li X, Nicklas RB. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–632. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

