Yeast and the AIDS virus: the odd couple
- PMID: 22778552
- PMCID: PMC3385842
- DOI: 10.1155/2012/549020
Yeast and the AIDS virus: the odd couple
Abstract
Despite being simple eukaryotic organisms, the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe have been widely used as a model to study human pathologies and the replication of human, animal, and plant viruses, as well as the function of individual viral proteins. The complete genome of S. cerevisiae was the first of eukaryotic origin to be sequenced and contains about 6,000 genes. More than 75% of the genes have an assigned function, while more than 40% share conserved sequences with known or predicted human genes. This strong homology has allowed the function of human orthologs to be unveiled starting from the data obtained in yeast. RNA plant viruses were the first to be studied in yeast. In this paper, we focus on the use of the yeast model to study the function of the proteins of human immunodeficiency virus type 1 (HIV-1) and the search for its cellular partners. This human retrovirus is the cause of AIDS. The WHO estimates that there are 33.4 million people worldwide living with HIV/AIDS, with 2.7 million new HIV infections per year and 2.0 million annual deaths due to AIDS. Current therapy is able to control the disease but there is no permanent cure or a vaccine. By using yeast, it is possible to dissect the function of some HIV-1 proteins and discover new cellular factors common to this simple cell and humans that may become potential therapeutic targets, leading to a long-lasting treatment for AIDS.
Figures
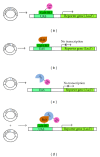



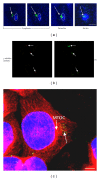


References
-
- Goffeau A, Barrell G, Bussey H, et al. Life with 6000 genes. Science. 1996;274(5287):546–567. - PubMed
-
- Mager WH, Winderickx J. Yeast as a model for medical and medicinal research. Trends in Pharmacological Sciences. 2005;26(5):265–273. - PubMed
-
- Steinmetz LM, Scharfe C, Deutschbauer AM, et al. Systematic screen for human disease genes in yeast. Nature Genetics. 2002;31(4):400–404. - PubMed
-
- Kushner DB, Lindenbach BD, Grdzelishvili VZ, Noueiry AO, Paul SM, Ahlquist P. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(26):15764–15769. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

