Three-dimensional expansion of a dynamic programming method for boundary detection and its application to sequential magnetic resonance imaging (MRI)
- PMID: 22778580
- PMCID: PMC3386679
- DOI: 10.3390/s120505195
Three-dimensional expansion of a dynamic programming method for boundary detection and its application to sequential magnetic resonance imaging (MRI)
Abstract
This study proposes a fast 3D dynamic programming expansion to find a shortest surface in a 3D matrix. This algorithm can detect boundaries in an image sequence. Using phantom image studies with added uniform distributed noise from different SNRs, the unsigned error of this proposed method is investigated. Comparing the automated results to the gold standard, the best averaged relative unsigned error of the proposed method is 0.77% (SNR = 20 dB), and its corresponding parameter values are reported. We further apply this method to detect the boundary of the real superficial femoral artery (SFA) in MRI sequences without a contrast injection. The manual tracings on the SFA boundaries are performed by well-trained experts to be the gold standard. The comparisons between the manual tracings and automated results are made on 16 MRI sequences (800 total images). The average unsigned error rate is 2.4% (SD = 2.0%). The results demonstrate that the proposed method can perform qualitatively better than the 2D dynamic programming for vessel boundary detection on MRI sequences.
Keywords: boundary detection; dynamic programming, MRI; femoral artery.
Figures
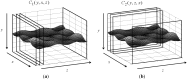



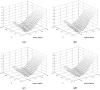




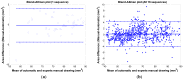

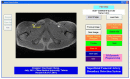
Similar articles
-
Accurate Measurement of Cross-Sectional Area of Femoral Artery on MRI Sequences of Transcontinental Ultramarathon Runners Using Optimal Parameters Selection.J Med Syst. 2016 Dec;40(12):260. doi: 10.1007/s10916-016-0626-y. Epub 2016 Oct 8. J Med Syst. 2016. PMID: 27722979
-
Automated localisation and boundary identification of superficial femoral artery on MRI sequences.Comput Methods Biomech Biomed Engin. 2013;16(8):873-84. doi: 10.1080/10255842.2011.643468. Epub 2012 Jan 6. Comput Methods Biomech Biomed Engin. 2013. PMID: 22220925
-
Automatic detection of the carotid artery boundary on cross-sectional MR image sequences using a circle model guided dynamic programming.Biomed Eng Online. 2011 Apr 11;10:26. doi: 10.1186/1475-925X-10-26. Biomed Eng Online. 2011. PMID: 21477378 Free PMC article.
-
Fast plaque burden assessment of the femoral artery using 3D black-blood MRI and automated segmentation.Med Phys. 2011 Oct;38(10):5370-84. doi: 10.1118/1.3633899. Med Phys. 2011. PMID: 21992357 Free PMC article.
-
Fast segmentation of the femoral arteries from 3D MR images: A tool for rapid assessment of peripheral arterial disease.Med Phys. 2015 May;42(5):2431-48. doi: 10.1118/1.4916803. Med Phys. 2015. PMID: 25979037
Cited by
-
Dynamic Programming Based Segmentation in Biomedical Imaging.Comput Struct Biotechnol J. 2017 Feb 16;15:255-264. doi: 10.1016/j.csbj.2017.02.001. eCollection 2017. Comput Struct Biotechnol J. 2017. PMID: 28289536 Free PMC article. Review.
-
Accurate Measurement of Cross-Sectional Area of Femoral Artery on MRI Sequences of Transcontinental Ultramarathon Runners Using Optimal Parameters Selection.J Med Syst. 2016 Dec;40(12):260. doi: 10.1007/s10916-016-0626-y. Epub 2016 Oct 8. J Med Syst. 2016. PMID: 27722979
-
In vivo microvascular network imaging of the human retina combined with an automatic three-dimensional segmentation method.J Biomed Opt. 2015 Jul;20(7):76003. doi: 10.1117/1.JBO.20.7.076003. J Biomed Opt. 2015. PMID: 26169790 Free PMC article.
-
Assessment of the endothelial function with changed volume of brachial artery by menstrual cycle.Biomed Eng Online. 2016 Sep 6;15(1):106. doi: 10.1186/s12938-016-0230-x. Biomed Eng Online. 2016. PMID: 27599988 Free PMC article.
References
-
- Ziou D., Tabbone S. Edge detection techniques: An overview. Int. J. Pattern Recogn. Image Anal. 1998;8:537–559.
-
- Zhai L., Dong S., Ma H. Recent Methods and Applications on Image Edge Detection. Proceedings of the 2008 International Workshop on Education Technology and Training & 2008 International Workshop on Geoscience and Remote Sensing; Shanghai, China. 21–22 December 2008.
-
- Kirbas C., Quek F. Vessel Extraction Techniques and Algorithms: A Survey. Proceedings of the 3rd IEEE Symposium on Bioinformatics and Bioengineering; Bethesda, MD, USA. 10–12 March 2003.
-
- Sonka M., Zhang X., Siebes M., Bissing M.S., Dejong S.C., Collins S.M., McKay C.R. Segmentation of intravascular ultrasound images: A knowledge-based approach. IEEE Trans. Med. Imaging. 1995;14:719–732. - PubMed
-
- Falcao A.X., Udupa J.K., Miyazawa F.K. An ultra-fast user-steered image segmentation paradigm: Live wire on the fly. IEEE Trans. Med. Imaging. 2000;19:55–62. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

