Y95 and E444 interaction required for high-affinity S-citalopram binding in the human serotonin transporter
- PMID: 22778858
- PMCID: PMC3369724
- DOI: 10.1021/cn100066p
Y95 and E444 interaction required for high-affinity S-citalopram binding in the human serotonin transporter
Abstract
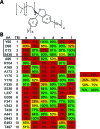
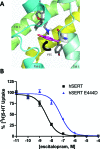
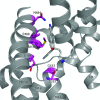
The human serotonin (5-hydroxytryptamine, 5-HT) transporter (hSERT) is responsible for the reuptake of 5-HT following synaptic release, as well as for import of the biogenic amine into several non-5-HT synthesizing cells including platelets. The antidepressant citalopram blocks SERT and thereby inhibits the transport of 5-HT. To identify key residues establishing high-affinity citalopram binding, we have built a comparative model of hSERT and Drosophila melanogaster SERT (dSERT) based on the Aquifex aeolicus leucine transporter (LeuT(Aa)) crystal structure. In this study, citalopram has been docked into the homology model of hSERT and dSERT using RosettaLigand. Our models reproduce the differential binding affinities for the R- and S-isomers of citalopram in hSERT and the impact of several hSERT mutants. Species-selective binding affinities for hSERT and dSERT also can be reproduced. Interestingly, the model predicts a hydrogen bond between E444 in transmembrane domain 8 (TM8) and Y95 in TM1 that places Y95 in a downward position, thereby removing Y95 from a direct interaction with S-citalopram. Mutation of E444D results in a 10-fold reduced binding affinity for S-citalopram, supporting the hypothesis that Y95 and E444 form a stabilizing interaction in the S-citalopram/hSERT complex.
Keywords: S-citalopram; computational docking; hSERT; homology model; ligand.
Figures
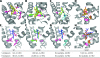
Similar articles
-
Allosteric effects of R- and S-citalopram on the human 5-HT transporter: evidence for distinct high- and low-affinity binding sites.Eur J Pharmacol. 2007 Jul 12;567(1-2):1-9. doi: 10.1016/j.ejphar.2007.03.055. Epub 2007 Apr 14. Eur J Pharmacol. 2007. PMID: 17499240
-
Mutational mapping and modeling of the binding site for (S)-citalopram in the human serotonin transporter.J Biol Chem. 2010 Jan 15;285(3):2051-63. doi: 10.1074/jbc.M109.072587. Epub 2009 Nov 5. J Biol Chem. 2010. PMID: 19892699 Free PMC article.
-
Comparative molecular field analysis using selectivity fields reveals residues in the third transmembrane helix of the serotonin transporter associated with substrate and antagonist recognition.J Pharmacol Exp Ther. 2008 Jun;325(3):791-800. doi: 10.1124/jpet.108.136200. Epub 2008 Mar 19. J Pharmacol Exp Ther. 2008. PMID: 18354055 Free PMC article.
-
Escitalopram, an antidepressant with an allosteric effect at the serotonin transporter--a review of current understanding of its mechanism of action.Psychopharmacology (Berl). 2012 Jan;219(1):1-13. doi: 10.1007/s00213-011-2463-5. Epub 2011 Sep 8. Psychopharmacology (Berl). 2012. PMID: 21901317 Review.
-
Three-dimensional models of neurotransmitter transporters and their interactions with cocaine and S-citalopram.World J Biol Psychiatry. 2006;7(2):99-109. doi: 10.1080/15622970500402144. World J Biol Psychiatry. 2006. PMID: 16684682 Review.
Cited by
-
Applications and Potential of In Silico Approaches for Psychedelic Chemistry.Molecules. 2023 Aug 9;28(16):5966. doi: 10.3390/molecules28165966. Molecules. 2023. PMID: 37630218 Free PMC article. Review.
-
(-)-Syringaresinol Exerts an Antidepressant-like Activity in Mice by Noncompetitive Inhibition of the Serotonin Transporter.Pharmaceuticals (Basel). 2024 Dec 5;17(12):1637. doi: 10.3390/ph17121637. Pharmaceuticals (Basel). 2024. PMID: 39770479 Free PMC article.
-
Insights to ligand binding to the monoamine transporters-from homology modeling to LeuBAT and dDAT.Front Pharmacol. 2015 Sep 24;6:208. doi: 10.3389/fphar.2015.00208. eCollection 2015. Front Pharmacol. 2015. PMID: 26441663 Free PMC article. Review.
-
Toward high-resolution modeling of small molecule-ion channel interactions.Front Pharmacol. 2024 Jun 11;15:1411428. doi: 10.3389/fphar.2024.1411428. eCollection 2024. Front Pharmacol. 2024. PMID: 38919257 Free PMC article.
-
Small-molecule ligand docking into comparative models with Rosetta.Nat Protoc. 2013;8(7):1277-98. doi: 10.1038/nprot.2013.074. Epub 2013 Jun 6. Nat Protoc. 2013. PMID: 23744289 Free PMC article.
References
-
-
http://www.who.int/mental_health/management/depression/definition/en/
-
-
- Hirschfeld R. M. (2000) J. Clin. Psychiatry. 61(Suppl. 6), 4–6. - PubMed
-
- Barker E. L.; Burris K. D.; Sanders-Bush E. (1991) Brain Res. 552, 330–332. - PubMed
-
- Blakely R. D.; Berson H. E.; Fremeau R. T. Jr.; Caron M. G.; Peek M. M.; Prince H. K.; Bradley C. C. (1991) Nature 354, 66–70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

