Disintegrins from hematophagous sources
- PMID: 22778902
- PMCID: PMC3386632
- DOI: 10.3390/toxins4050296
Disintegrins from hematophagous sources
Abstract
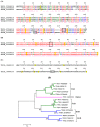
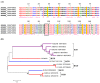
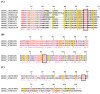
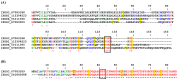
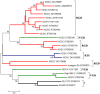
Bloodsucking arthropods are a rich source of salivary molecules (sialogenins) which inhibit platelet aggregation, neutrophil function and angiogenesis. Here we review the literature on salivary disintegrins and their targets. Disintegrins were first discovered in snake venoms, and were instrumental in our understanding of integrin function and also for the development of anti-thrombotic drugs. In hematophagous animals, most disintegrins described so far have been discovered in the salivary gland of ticks and leeches. A limited number have also been found in hookworms and horseflies, and none identified in mosquitoes or sand flies. The vast majority of salivary disintegrins reported display a RGD motif and were described as platelet aggregation inhibitors, and few others as negative modulator of neutrophil or endothelial cell functions. This notably low number of reported disintegrins is certainly an underestimation of the actual complexity of this family of proteins in hematophagous secretions. Therefore an algorithm was created in order to identify the tripeptide motifs RGD, KGD, VGD, MLD, KTS, RTS, WGD, or RED (flanked by cysteines) in sialogenins deposited in GenBank database. The search included sequences from various blood-sucking animals such as ticks (e.g., Ixodes sp., Argas sp., Rhipicephalus sp., Amblyommasp.), tabanids (e.g., Tabanus sp.), bugs (e.g., Triatoma sp., Rhodnius prolixus), mosquitoes (e.g., Anopheles sp., Aedes sp., Culex sp.), sand flies (e.g., Lutzomyia sp., Phlebotomus sp.), leeches (e.g., Macrobdella sp., Placobdella sp.) and worms (e.g., Ancylostoma sp.). This approach allowed the identification of a remarkably high number of novel putative sialogenins with tripeptide motifs typical of disintegrins (>450 sequences) whose biological activity remains to be verified. This database is accessible online as a hyperlinked worksheet and displays biochemical, taxonomic, and gene ontology aspects for each putative disintegrin. It is also freely available for download (right click with the mouse) at links http://exon.niaid.nih.gov/transcriptome/RGD/RGD-Peps-WEB.xlsx (web version) and http://exon.niaid.nih.gov/transcriptome/RGD/RGD-sialogenins.zip (stand alone version).
Keywords: angiogenesis; bloodsucking; disintegrins; hematophagy; platelet aggregation; proteome; salivary; sialogenins; sialome; snake venom; thrombus; transcriptome.
Figures



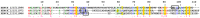
References
-
- Hood J.D., Cheresh D.A. Role of integrins in cell invasion and migration. Nat. Rev. Cancer. 2002;2:91–100. - PubMed
-
- Silva R., D’Amico G., Hodivala-Dilke K.M., Reynolds L.E. Integrins: The keys to unlocking angiogenesis. Arterioscler. Thromb. Vasc. Biol. 2008;28:1703–1713. - PubMed
-
- McLane M.A., Joerger T., Mahmoud A. Disintegrins in health and disease. Front Biosci. 2008;13:6617–6637. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

