DNA methylation and its basic function
- PMID: 22781841
- PMCID: PMC3521964
- DOI: 10.1038/npp.2012.112
DNA methylation and its basic function
Abstract
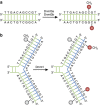
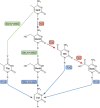
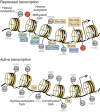
In the mammalian genome, DNA methylation is an epigenetic mechanism involving the transfer of a methyl group onto the C5 position of the cytosine to form 5-methylcytosine. DNA methylation regulates gene expression by recruiting proteins involved in gene repression or by inhibiting the binding of transcription factor(s) to DNA. During development, the pattern of DNA methylation in the genome changes as a result of a dynamic process involving both de novo DNA methylation and demethylation. As a consequence, differentiated cells develop a stable and unique DNA methylation pattern that regulates tissue-specific gene transcription. In this chapter, we will review the process of DNA methylation and demethylation in the nervous system. We will describe the DNA (de)methylation machinery and its association with other epigenetic mechanisms such as histone modifications and noncoding RNAs. Intriguingly, postmitotic neurons still express DNA methyltransferases and components involved in DNA demethylation. Moreover, neuronal activity can modulate their pattern of DNA methylation in response to physiological and environmental stimuli. The precise regulation of DNA methylation is essential for normal cognitive function. Indeed, when DNA methylation is altered as a result of developmental mutations or environmental risk factors, such as drug exposure and neural injury, mental impairment is a common side effect. The investigation into DNA methylation continues to show a rich and complex picture about epigenetic gene regulation in the central nervous system and provides possible therapeutic targets for the treatment of neuropsychiatric disorders.
Figures
References
-
- Aapola U, Kawasaki K, Scott HS, Ollila J, Vihinen M, Heino M, et al. 2000Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family Genomics 65293–298.Identified Dnmt3L and profiled its expression. - PubMed
-
- Aapola U, Lyle R, Krohn K, Antonarakis SE, Peterson P. Isolation and initial characterization of the mouse Dnmt3l gene. Cytogenet Cell Genet. 2001;92:122–126. - PubMed
-
- Achour M, Jacq X, Ronde P, Alhosin M, Charlot C, Chataigneau T, et al. The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene. 2008;27:2187–2197. - PubMed
-
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY.1999Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2 Nat Genet 23185–188.Discovered MECP2 mutation as the cause of Rett Syndrome and paved way for further study of MECP2 protein role in the nervous system. - PubMed
-
- Aran D, Toperoff G, Rosenberg M, Hellman A. Replication timing-related and gene body-specific methylation of active human genes. Hum Mol Genet. 2011;20:670–680. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

