Diversity and subcellular distribution of archaeal secreted proteins
- PMID: 22783239
- PMCID: PMC3387779
- DOI: 10.3389/fmicb.2012.00207
Diversity and subcellular distribution of archaeal secreted proteins
Abstract
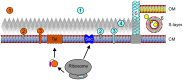
Secreted proteins make up a significant percentage of a prokaryotic proteome and play critical roles in important cellular processes such as polymer degradation, nutrient uptake, signal transduction, cell wall biosynthesis, and motility. The majority of archaeal proteins are believed to be secreted either in an unfolded conformation via the universally conserved Sec pathway or in a folded conformation via the Twin arginine transport (Tat) pathway. Extensive in vivo and in silico analyses of N-terminal signal peptides that target proteins to these pathways have led to the development of computational tools that not only predict Sec and Tat substrates with high accuracy but also provide information about signal peptide processing and targeting. Predictions therefore include indications as to whether a substrate is a soluble secreted protein, a membrane or cell wall anchored protein, or a surface structure subunit, and whether it is targeted for post-translational modification such as glycosylation or the addition of a lipid. The use of these in silico tools, in combination with biochemical and genetic analyses of transport pathways and their substrates, has resulted in improved predictions of the subcellular localization of archaeal secreted proteins, allowing for a more accurate annotation of archaeal proteomes, and has led to the identification of potential adaptations to extreme environments, as well as phyla-specific pathways among the archaea. A more comprehensive understanding of the transport pathways used and post-translational modifications of secreted archaeal proteins will also facilitate the identification and heterologous expression of commercially valuable archaeal enzymes.
Keywords: Sec transport; Tat transport; archaea; archaeosortase; cell surface structures; lipoprotein; pili; protein transport.
Figures
Similar articles
-
ArtA-Dependent Processing of a Tat Substrate Containing a Conserved Tripartite Structure That Is Not Localized at the C Terminus.J Bacteriol. 2017 Mar 14;199(7):e00802-16. doi: 10.1128/JB.00802-16. Print 2017 Apr 1. J Bacteriol. 2017. PMID: 28069824 Free PMC article.
-
Haloferax volcanii twin-arginine translocation substates include secreted soluble, C-terminally anchored and lipoproteins.Mol Microbiol. 2007 Dec;66(6):1597-606. doi: 10.1111/j.1365-2958.2007.06034.x. Mol Microbiol. 2007. PMID: 18045386
-
Signal Peptide Hydrophobicity Modulates Interaction with the Twin-Arginine Translocase.mBio. 2017 Aug 1;8(4):e00909-17. doi: 10.1128/mBio.00909-17. mBio. 2017. PMID: 28765221 Free PMC article.
-
Protein transport in Archaea: Sec and twin arginine translocation pathways.Curr Opin Microbiol. 2005 Dec;8(6):713-9. doi: 10.1016/j.mib.2005.10.006. Epub 2005 Oct 27. Curr Opin Microbiol. 2005. PMID: 16257258 Review.
-
The archaeal twin-arginine translocation pathway.Biochem Soc Trans. 2003 Jun;31(Pt 3):686-9. doi: 10.1042/bst0310686. Biochem Soc Trans. 2003. PMID: 12773183 Review.
Cited by
-
S-layers: principles and applications.FEMS Microbiol Rev. 2014 Sep;38(5):823-64. doi: 10.1111/1574-6976.12063. Epub 2014 Feb 24. FEMS Microbiol Rev. 2014. PMID: 24483139 Free PMC article.
-
Molecular characterization of the thioredoxin system from Methanosarcina acetivorans.FEBS J. 2014 Oct;281(20):4598-611. doi: 10.1111/febs.12964. Epub 2014 Sep 6. FEBS J. 2014. PMID: 25112424 Free PMC article.
-
On the Archaeal Origins of Eukaryotes and the Challenges of Inferring Phenotype from Genotype.Trends Cell Biol. 2016 Jul;26(7):476-485. doi: 10.1016/j.tcb.2016.03.009. Epub 2016 Jun 16. Trends Cell Biol. 2016. PMID: 27319280 Free PMC article. Review.
-
The modern "3G" age of archaeal molecular biology.Front Microbiol. 2012 Dec 20;3:430. doi: 10.3389/fmicb.2012.00430. eCollection 2012. Front Microbiol. 2012. PMID: 23267357 Free PMC article. No abstract available.
-
Biomimetic interfaces based on S-layer proteins, lipid membranes and functional biomolecules.J R Soc Interface. 2014 May 8;11(96):20140232. doi: 10.1098/rsif.2014.0232. Print 2014 Jul 6. J R Soc Interface. 2014. PMID: 24812051 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

