Inhibition of methane oxidation by nitrogenous fertilizers in a paddy soil
- PMID: 22783249
- PMCID: PMC3389332
- DOI: 10.3389/fmicb.2012.00246
Inhibition of methane oxidation by nitrogenous fertilizers in a paddy soil
Abstract
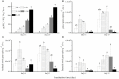
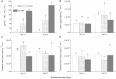
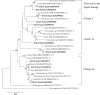
Nitrogenous fertilizers are generally thought to have an important role in regulating methane oxidation. In this study, the effect of ammonium on methane oxidation activity was investigated in a paddy soil using urea at concentrations of 0, 50, 100, 200, and 400 μg N per gram dry weight soil (N/g.d.w.s) and ammonium sulfate at concentrations of 0, 50, and 200 μg N/g.d.w.s. The results of this study demonstrate that urea concentrations of 200 μg N/g.d.w.s. and above significantly inhibit methane oxidation activity, whereas no statistically significant difference was observed in methane oxidation activity among soil microcosms with urea concentrations of less than 200 μg N/g.d.w.s after incubation for 27 days. Similar results were obtained in a sense that methane oxidation activity was inhibited only when the ammonium sulfate concentration was 200 μg N/g.d.w.s in soil microcosms in this study. Phylogenetic analysis of pmoA genes showed that nitrogen fertilization resulted in apparent changes in the community composition of methane-oxidizing bacteria (MOB). Type I MOB displayed an increased abundance in soil microcosms amended with nitrogenous fertilizers, whereas type II MOB dominated the native soil. Furthermore, although no statistically significant relationship was observed between pmoA gene and amoA gene abundances, methane oxidation activity was significantly negatively correlated with nitrification activity in the presence of urea or ammonium sulfate. Our results indicate that the methane oxidation activity in paddy soils might be inhibited when the concentration of ammonium fertilizers is high and that the interactions between ammonia and methane oxidizers need to be further investigated.
Keywords: methane oxidation; nitrification activity; nitrogenous fertilizers; paddy soil; particulate methane monooxygenase gene pmoa.
Figures






References
-
- Acton S. D., Baggs E. M. (2011). Interactions between N application rate, CH4 oxidation and N2O production in soil. Biogeochemistry 103, 15–26
-
- Aronson E. L., Helliker B. R. (2010). Methane flux in non-wetland soils in response to nitrogen addition: a meta-analysis. Ecology 91, 3242–3251 - PubMed
LinkOut - more resources
Full Text Sources

