Chloroplast envelope protein targeting fidelity is independent of cytosolic components in dual organelle assays
- PMID: 22783268
- PMCID: PMC3384937
- DOI: 10.3389/fpls.2012.00148
Chloroplast envelope protein targeting fidelity is independent of cytosolic components in dual organelle assays
Abstract
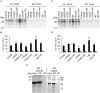
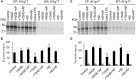
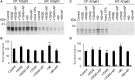
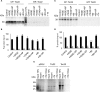
The general mechanisms of intracellular protein targeting are well established, and depend on a targeting sequence in the protein, which is recognized by a targeting factor. Once a membrane protein is delivered to the correct organelle its targeting sequence can be recognized by receptors and a translocase, leading to membrane insertion. However, the relative contribution of each step for generating fidelity and efficiency of the overall process has not been systematically addressed. Here, we use tail-anchored (TA) membrane proteins in cell-free competitive targeting assays to chloroplasts to show that targeting can occur efficiently and with high fidelity in the absence of all cytosolic components, suggesting that chloroplast envelope protein targeting is primarily dependent on events at the outer envelope. Efficiency of targeting was increased by the addition of complete cytosol, and by Hsp70 or Hsp90, depending on the protein, but none of these cytosolic components influenced the fidelity of targeting. Our results suggest that the main role of targeting factors in chloroplast localization is to increase targeting efficiency by maintaining recognition competency at the outer envelope.
Keywords: competitive targeting; molecular chaperones; protein localization; tail-anchored protein; targeting factors.
Figures


Similar articles
-
Biogenesis of chloroplast outer envelope membrane proteins.Plant Cell Rep. 2019 Jul;38(7):783-792. doi: 10.1007/s00299-019-02381-6. Epub 2019 Jan 22. Plant Cell Rep. 2019. PMID: 30671649 Review.
-
OEP61 is a chaperone receptor at the plastid outer envelope.Biochem J. 2011 Aug 15;438(1):143-53. doi: 10.1042/BJ20110448. Biochem J. 2011. PMID: 21612577 Free PMC article.
-
Distinct pathways mediate the sorting of tail-anchored proteins to the plastid outer envelope.PLoS One. 2010 Apr 14;5(4):e10098. doi: 10.1371/journal.pone.0010098. PLoS One. 2010. PMID: 20418952 Free PMC article.
-
New Insights into the Chloroplast Outer Membrane Proteome and Associated Targeting Pathways.Int J Mol Sci. 2022 Jan 29;23(3):1571. doi: 10.3390/ijms23031571. Int J Mol Sci. 2022. PMID: 35163495 Free PMC article. Review.
-
Membrane-Specific Targeting of Tail-Anchored Proteins SECE1 and SECE2 Within Chloroplasts.Front Plant Sci. 2019 Nov 8;10:1401. doi: 10.3389/fpls.2019.01401. eCollection 2019. Front Plant Sci. 2019. PMID: 31781139 Free PMC article.
Cited by
-
Exploring mechanisms linked to differentiation and function of dimorphic chloroplasts in the single cell C4 species Bienertia sinuspersici.BMC Plant Biol. 2014 Jan 21;14:34. doi: 10.1186/1471-2229-14-34. BMC Plant Biol. 2014. PMID: 24443986 Free PMC article.
-
Specific targeting of proteins to outer envelope membranes of endosymbiotic organelles, chloroplasts, and mitochondria.Front Plant Sci. 2014 Apr 29;5:173. doi: 10.3389/fpls.2014.00173. eCollection 2014. Front Plant Sci. 2014. PMID: 24808904 Free PMC article. Review.
-
Biogenesis of chloroplast outer envelope membrane proteins.Plant Cell Rep. 2019 Jul;38(7):783-792. doi: 10.1007/s00299-019-02381-6. Epub 2019 Jan 22. Plant Cell Rep. 2019. PMID: 30671649 Review.
-
New insights into the targeting of a subset of tail-anchored proteins to the outer mitochondrial membrane.Front Plant Sci. 2014 Sep 4;5:426. doi: 10.3389/fpls.2014.00426. eCollection 2014. Front Plant Sci. 2014. PMID: 25237314 Free PMC article.
-
Go your own way: membrane-targeting sequences.Plant Physiol. 2021 Apr 2;185(3):608-618. doi: 10.1093/plphys/kiaa058. Plant Physiol. 2021. PMID: 33822216 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

