Transcriptional interference networks coordinate the expression of functionally related genes clustered in the same genomic loci
- PMID: 22783276
- PMCID: PMC3389743
- DOI: 10.3389/fgene.2012.00122
Transcriptional interference networks coordinate the expression of functionally related genes clustered in the same genomic loci
Abstract
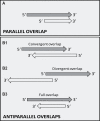
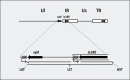
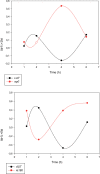
The regulation of gene expression is essential for normal functioning of biological systems in every form of life. Gene expression is primarily controlled at the level of transcription, especially at the phase of initiation. Non-coding RNAs are one of the major players at every level of genetic regulation, including the control of chromatin organization, transcription, various post-transcriptional processes, and translation. In this study, the Transcriptional Interference Network (TIN) hypothesis was put forward in an attempt to explain the global expression of antisense RNAs and the overall occurrence of tandem gene clusters in the genomes of various biological systems ranging from viruses to mammalian cells. The TIN hypothesis suggests the existence of a novel layer of genetic regulation, based on the interactions between the transcriptional machineries of neighboring genes at their overlapping regions, which are assumed to play a fundamental role in coordinating gene expression within a cluster of functionally linked genes. It is claimed that the transcriptional overlaps between adjacent genes are much more widespread in genomes than is thought today. The Waterfall model of the TIN hypothesis postulates a unidirectional effect of upstream genes on the transcription of downstream genes within a cluster of tandemly arrayed genes, while the Seesaw model proposes a mutual interdependence of gene expression between the oppositely oriented genes. The TIN represents an auto-regulatory system with an exquisitely timed and highly synchronized cascade of gene expression in functionally linked genes located in close physical proximity to each other. In this study, we focused on herpesviruses. The reason for this lies in the compressed nature of viral genes, which allows a tight regulation and an easier investigation of the transcriptional interactions between genes. However, I believe that the same or similar principles can be applied to cellular organisms too.
Keywords: Hox genes; antisense RNA; genomic organization; herpesvirus; polycistronic RNAs; pseudorabies virus; tandem genes; transcriptional interference.
Figures




Similar articles
-
Full-Length Isoform Sequencing Reveals Novel Transcripts and Substantial Transcriptional Overlaps in a Herpesvirus.PLoS One. 2016 Sep 29;11(9):e0162868. doi: 10.1371/journal.pone.0162868. eCollection 2016. PLoS One. 2016. PMID: 27685795 Free PMC article.
-
Functionally Related Genes Cluster into Genomic Regions That Coordinate Transcription at a Distance in Saccharomyces cerevisiae.mSphere. 2019 Mar 13;4(2):e00063-19. doi: 10.1128/mSphere.00063-19. mSphere. 2019. PMID: 30867326 Free PMC article.
-
Interactions between the transcription and replication machineries regulate the RNA and DNA synthesis in the herpesviruses.Virus Genes. 2019 Jun;55(3):274-279. doi: 10.1007/s11262-019-01643-5. Epub 2019 Feb 14. Virus Genes. 2019. PMID: 30767118 Free PMC article. Review.
-
Noncontiguous operon is a genetic organization for coordinating bacterial gene expression.Proc Natl Acad Sci U S A. 2019 Jan 29;116(5):1733-1738. doi: 10.1073/pnas.1812746116. Epub 2019 Jan 11. Proc Natl Acad Sci U S A. 2019. PMID: 30635413 Free PMC article.
-
Post-transcriptional regulation of polycistronic microRNAs.Wiley Interdiscip Rev RNA. 2023 Mar;14(2):e1749. doi: 10.1002/wrna.1749. Epub 2022 Jun 14. Wiley Interdiscip Rev RNA. 2023. PMID: 35702737 Review.
Cited by
-
Full-Length Isoform Sequencing Reveals Novel Transcripts and Substantial Transcriptional Overlaps in a Herpesvirus.PLoS One. 2016 Sep 29;11(9):e0162868. doi: 10.1371/journal.pone.0162868. eCollection 2016. PLoS One. 2016. PMID: 27685795 Free PMC article.
-
Multi-Platform Sequencing Approach Reveals a Novel Transcriptome Profile in Pseudorabies Virus.Front Microbiol. 2018 Jan 22;8:2708. doi: 10.3389/fmicb.2017.02708. eCollection 2017. Front Microbiol. 2018. PMID: 29403453 Free PMC article.
-
Combined Short and Long-Read Sequencing Reveals a Complex Transcriptomic Architecture of African Swine Fever Virus.Viruses. 2021 Mar 30;13(4):579. doi: 10.3390/v13040579. Viruses. 2021. PMID: 33808073 Free PMC article.
-
Integrative profiling of Epstein-Barr virus transcriptome using a multiplatform approach.Virol J. 2022 Jan 6;19(1):7. doi: 10.1186/s12985-021-01734-6. Virol J. 2022. PMID: 34991630 Free PMC article.
-
Long-read transcriptomics of caviid gammaherpesvirus 1: compiling a comprehensive RNA atlas.mSystems. 2025 Mar 18;10(3):e0167824. doi: 10.1128/msystems.01678-24. Epub 2025 Feb 27. mSystems. 2025. PMID: 40013795 Free PMC article.
References
-
- Audic S., Claverie J. M. (1997). The significance of digital gene expression profiles. Genome Res. 7 986–995 - PubMed
-
- Bertone P., Stolc V., Royce T. E., Rozowsky J. S., Urban A. E., Zhu X., Rinn J. L., Tongprasit W., Samanta M., Weissman S., Gerstein M., Snyder M. (2004). Global identification of human transcribed sequences with genome tiling arrays. Science 306 2242–2246 - PubMed
-
- Birney E., Stamatoyannopoulos J. A., Dutta A., Guigo R., Gingeras T. R., Margulies E. H., Weng Z., Snyder M., Dermitzakis E. T., Thurman R. E., Kuehn M. S., Taylor C. M., Neph S., Koch C. M., Asthana S., Malhotra A., Adzhubei I., Greenbaum J. A., Andrews R. M., Flicek P., Boyle P. J., Cao H., Carter N. P., Clelland G. K., Davis S., Day N., Dhami P., Dillon S. C., Dorschner M. O., Fiegler H., Giresi P. G., Goldy J., Hawrylycz M., Haydock A., Humbert R., James K. D., Johnson B. E., Johnson E. M., Frum T. T., Rosenzweig E. R., Karnani N., Lee K., Lefebvre G. C., Navas P. A., Neri F., Parker S. C., Sabo P. J., Sandstrom R., Shafer A., Vetrie D., Weaver M., Wilcox S., Yu M., Collins F. S., Dekker J., Lieb J. D., Tullius T. D., Crawford G. E., Sunyaev S., Noble W. S., Dunham I., Denoeud F., Reymond A., Kapranov P., Rozowsky J., Zheng D., Castelo R., Frankish A., Harrow J., Ghosh S., Sandelin A., Hofacker I. L., Baertsch R., Keefe D., Dike S., Cheng J., Hirsch H. A., Sekinger E. A., Lagarde J., Abril J. F., Shahab A., Flamm C., Fried C., Hackermüller J., Hertel J., Lindemeyer M., Missal K., Tanzer A., Washietl S., Korbel J., Emanuelsson O., Pedersen J. S., Holroyd N., Taylor R., Swarbreck D., Matthews N., Dickson M. C., Thomas D. J., Weirauch M. T., Gilbert J., Drenkow J., Bell I., Zhao X., Srinivasan K. G., Sung W. K., Ooi H. S., Chiu K. P., Foissac S., Alioto T., Brent M., Pachter L., Tress M. L., Valencia A., Choo S. W., Choo C. Y., Ucla C., Manzano C., Wyss C., Cheung E., Clark T. G., Brown J. B., Ganesh M., Patel S., Tammana H., Chrast J., Henrichsen C. N., Kai C., Kawai J., Nagalakshmi U., Wu J., Lian Z., Lian J., Newburger P., Zhang X., Bickel P., Mattick J. S., Carninci P., Hayashizaki Y., Weissman S., Hubbard T., Myers R. M., Rogers J., Stadler P. F., Lowe T. M., Wei C. L., Ruan Y., Struhl K., Gerstein M., Antonarakis S. E., Fu Y., Green E. D., Karaöz U., Siepel A., Taylor J., Liefer L. A., Wetterstrand K. A., Good P. J., Feingold E. A., Guyer M. S., Cooper G. M., Asimenos G., Dewey C. N., Hou M., Nikolaev S., Montoya-Burgos J. I., Löytynoja A., Whelan S., Pardi F., Massingham T., Huang H., Zhang N. R., Holmes I., Mullikin J. C., Ureta-Vidal A., Paten B., Seringhaus M., Church D., Rosenbloom K., Kent W. J., Stone E. A., NISC Comparative Sequencing Program; Baylor College of Medicine Human Genome Sequencing Center; Washington University Genome Sequencing Center; Broad Institute; Children’s Hospital Oakland Research Institute; Batzoglou S., Goldman N., Hardison R. C., Haussler D., Miller W., Sidow A., Trinklein N. D., Zhang Z. D., Barrera L., Stuart R., King D. C., Ameur A., Enroth S., Bieda M. C., Kim J., Bhinge A. A., Jiang N., Liu J., Yao F., Vega V. B., Lee C. W., Ng P., Shahab A., Yang A., Moqtaderi Z., Zhu Z., Xu X., Squazzo S., Oberley M. J., Inman D., Singer M. A., Richmond T. A., Munn K. J., Rada-Iglesias A., Wallerman O., Komorowski J., Fowler J. C., Couttet P., Bruce A. W., Dovey O. M., Ellis P. D., Langford C. F., Nix D. A., Euskirchen G., Hartman S., Urban A. E., Kraus P., Van Calcar S., Heintzman N., Kim T. H., Wang K., Qu C., Hon G., Luna R., Glass C. K., Rosenfeld M. G., Aldred S. F., Cooper S. J., Halees A., Lin J. M., Shulha H. P., Zhang X., Xu M., Haidar J. N., Yu Y., Ruan Y., Iyer V. R., Green R. D., Wadelius C., Farnham P. J., Ren B., Harte R. A., Hinrichs A. S., Trumbower H., Clawson H., Hillman-Jackson J., Zweig A. S., Smith K., Thakkapallayil A., Barber G., Kuhn R. M., Karolchik D., Armengol L., Bird C. P., de Bakker P. I., Kern A. D., Lopez-Bigas N., Martin J. D., Stranger B. E., Woodroffe A., Davydov E., Dimas A., Eyras E., Hallgrímsdóttir I. B., Huppert J., Zody M. C., Abecasis G. R., Estivill X., Bouffard G. G., Guan X., Hansen N. F., Idol J. R., Maduro V. V., Maskeri B., McDowell J. C., Park M., Thomas P. J., Young A. C., Blakesley R. W., Muzny D. M., Sodergren E., Wheeler D. A., Worley K. C., Jiang H., Weinstock G. M., Gibbs R. A., Graves T., Fulton R., Mardis E. R., Wilson R. K., Clamp M., Cuff J., Gnerre S., Jaffe D. B., Chang J. L., Lindblad-Toh K., Lander E. S., Koriabine M., Nefedov M., Osoegawa K., Yoshinaga Y., Zhu B., de Jong P. J. (2007). Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447 799–816 - PMC - PubMed
LinkOut - more resources
Full Text Sources

