microRNA-103 regulates the growth and invasion of endometrial cancer cells through the downregulation of tissue inhibitor of metalloproteinase 3
- PMID: 22783422
- PMCID: PMC3392565
- DOI: 10.3892/ol.2012.638
microRNA-103 regulates the growth and invasion of endometrial cancer cells through the downregulation of tissue inhibitor of metalloproteinase 3
Abstract
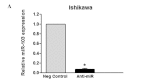
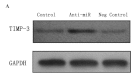
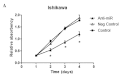
Despite improvements in treatment over the past few decades, endometrial cancer remains one of the most common causes of mortality in women and there is an urgent need for the development of targeted therapies. The aim of this study was to confirm the target gene of miR-103 in human endometrial cancer and investigate the biological functions in which miR-103 is involved through the regulation of the expression of its target gene. This study may provide useful data to gain a better understanding of the effect of miR-103 in tumor formation. miR-103 expression levels were measured using real-time quantitative PCR. The effect of miR-103 on tissue inhibitor of metalloproteinase 3 (TIMP-3) expression was assessed in endometrial cancer cell lines with a miR-103 inhibitor to decrease the level of miR-103 expression. Furthermore, the roles of miR-103 in cell growth and invasion were analyzed using miR-103 inhibitor-transfected cells. The level of expression of miR-103 decreased following transfection with the miR-103 inhibitor. miR-103 inhibitor transfection increased the activity of the luciferase reporter assay containing the TIMP-3 3'-untranslated region (UTR) construct and increased the levels of the TIMP-3 protein but not its mRNA in endometrial cancer cell lines. Finally, miR-103 inhibitor-transfected cells exhibited reduced cell growth and invasive characteristics. Our data suggested that miR-103 post-transcriptionally downregulates the expression of the tumor suppressor TIMP-3 and stimulates growth and invasion in endometrial cancer cell lines. This provides a possible therapeutic target that may upregulate TIMP-3 in endometrial cancer.
Figures









References
-
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. - PubMed
-
- Xi Y, Shalgi R, Fodstad O, Pilpel Y, Ju J. Differentially regulated micro-RNAs and actively translated messenger RNA transcripts by tumor suppressor p53 in colon cancer. Clin Cancer Res. 2006;12:2014–2024. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
