Structural analysis of B-cell epitopes in antibody:protein complexes
- PMID: 22784991
- PMCID: PMC3461403
- DOI: 10.1016/j.molimm.2012.06.001
Structural analysis of B-cell epitopes in antibody:protein complexes
Abstract
The binding of antigens to antibodies is one of the key events in an immune response against foreign molecules and is a critical element of several biomedical applications including vaccines and immunotherapeutics. For development of such applications, the identification of antibody binding sites (B-cell epitopes) is essential. However experimental epitope mapping is highly cost-intensive and computer-aided methods do in general have moderate performance. One major reason for this moderate performance is an incomplete understanding of what characterizes an epitope. To fill this gap, we here developed a novel framework for comparing and superimposing B-cell epitopes and applied it on a dataset of 107 non-similar antigen:antibody structures extracted from the PDB database. With the presented framework, we were able to describe the general B-cell epitope as a flat, oblong, oval shaped volume consisting of predominantly hydrophobic amino acids in the center flanked by charged residues. The average epitope was found to be made up of ∼15 residues with one linear stretch of 5 or more residues constituting more than half of the epitope size. Furthermore, the epitope area is predominantly constrained to a plane above the antibody tip, in which the epitope is orientated in a -30° to 60° angle relative to the light to heavy chain antibody direction. Contrary to previously findings, we did not find a significant deviation between the amino acid composition in epitopes and the composition of equally exposed parts of the antigen surface. Our results, in combination with previously findings, give a detailed picture of the B-cell epitope that may be used in development of improved B-cell prediction methods.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures


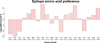

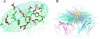


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

